Data Source Lightweight Integration (version alpha 0.1)Deprecated: Use the jsCDK insteadGenomeSpace Data Source Lightweight IntegrationMost tools in the GenomeSpace tool bar are integrated with GenomeSpace using either the GenomeSpace API or the GenomeSpace Java Client Development Kit (CDK). One of these two approaches is required to achieve full integration, meaning that users have integrated identity (single sign on) and can send data to the tool from GenomeSpace, to GenomeSpace from the tool, and possibly also directly launch other GenomeSpace tools. For a subset of applications however, such as data portals, the requirements are more modest, and so they only need to be able to send data into GenomeSpace for analysis. Data portals have data to share for analysis in GenomeSpace tools, but do not have the capability to take data back from GenomeSpace or analyze it themselves. For tools that fall into this category, we have developed the GenomeSpace Lightweight Data Source Integration JavaScript Library (DSIL). FeaturesThe DSIL allows web sites to:
The DSIL handles all aspects of GenomeSpace authentication and provides a user interface that allows a user:
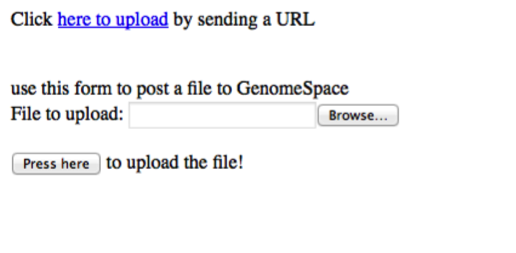
In short, it acts as a web-based save-file dialog that accepts either a URL or a posted file. The DSIL is implemented in two parts: first as a JavaScript library hosted by the GenomeSpace user interface, and second as a web page that handles authentication, GenomeSpace file system navigation, and file (re)naming. Example Use of the DSILAn example page is available for live testing at http://gsui.genomespace.org/jsui/jsCDK/Demo.html. This simple example page looks like this;
Clicking on the here to upload link or adding a file and clicking the button will open a new browser window. The new window will authenticate with GenomeSpace and then present the user with the ability to select the target directory and filename to use.
The full source of the page is as follows: <html><head> <meta http-equiv="Content-Type" content="text/html; charset=UTF-8"> <title>Upload To GenomeSpace Example</title> <script src="https://gsui.genomespace.org/jsui/js/jquery-1.7.2.min.js" type="text/javascript"></script> <script src="https://gsui.genomespace.org/jsui/upload/gsuploadwindow.js" type="text/javascript"></script> </head> <body> <div id="uploadLink">Click <a href="#" onclick="uploadUrlByGetToGS();return false;">here to upload</a> by sending a URL</div> <br><br> use this form to post a file to GenomeSpace <form method="POST" enctype="multipart/form-data" id="postToGenomeSpace"> File to upload: <input name="upfile" type="file"><br/><br/> <input id="postUploadBtn" value="Press here" onclick="postUploadToGS(); return false;" type="button"> to upload the file! </form> <script type="text/javascript"> function uploadUrlByGetToGS(){ var urlToUpload = "http://beta.flymine.org/beta/service/query/results?format=tab&start=0&query=%3Cquery+model%3D%22genomic%22+view%3D%22Gene.symbol+Gene.organism.name+Gene.chromosomeLocation.locatedOn.primaryIdentifier+Gene.chromosomeLocation.start+Gene.chromosomeLocation.end+Gene.proteins.name+Gene.exons.primaryIdentifier%22+%3E%3Cconstraint+path%3D%22Gene.chromosomeLocation.locatedOn%22+type%3D%22Chromosome%22+%2F%3E%3Cconstraint+path%3D%22Gene.length%22+op%3D%22%26lt%3B%22+value%3D%228000%22+%2F%3E%3Cconstraint+path%3D%22Gene.pathways.name%22+op%3D%22ONE+OF%22+%3E%3Cvalue%3EMetabolic+pathways%3C%2Fvalue%3E%3Cvalue%3EGene+Expression%3C%2Fvalue%3E%3Cvalue%3ETranscription%3C%2Fvalue%3E%3Cvalue%3EmRNA+Processing%3C%2Fvalue%3E%3C%2Fconstraint%3E%3Cconstraint+path%3D%22Gene.chromosome.primaryIdentifier%22+op%3D%22%3D%22+value%3D%222L%22+%2F%3E%3C%2Fquery%3E&_=1362507."; var onComplete = function(savePath){ alert('outer Saved to GenomeSpace as ' + savePath); }; var onError = function(savePath){ alert('outer ERROR saving to GenomeSpace as ' + savePath); }; gsUploadByGet({ url: urlToUpload, filename: 'TheFileName.txt', successCallback: onComplete, errorCallback: onError }); return false; }; function postUploadToGS(){ var formData = new FormData($('#postToGenomeSpace')[0]); gsUploadByPost(formData); return false; }; </script> </body></html> The basic example above consists of
Let's look at each part in turn. Included JavaScript LibrariesTwo JavaScript libraries are used for this example: <script src="https://gsui.genomespace.org/jsui/js/jquery-1.7.2.min.js" type="text/javascript"></script> <script src="https://gsui.genomespace.org/jsui/upload/gsuploadwindow.js" type="text/javascript"></script> The JQuery library (loaded directly from the GenomeSpace User Interface site) is required only for doing the POST version (more details below). The gsuploadwindow.js library has the JavaScript methods to handle the GET/POST of files to GenomeSpace. Sending a File via a GETTo send a file to GenomeSpace that it must GET from a server, the URL sent must be a publicly accessible URL, since the DSIL assumes that the sending web application is not using GenomeSpace Single Sign On. In this mode, the GenomeSpace server will use an HTTP GET to retrieve the file contents. Since this is being done by the GenomeSpace server and not the user's browser, there will be no authentication information, headers, or cookies passed in the request. Sending a URL for GenomeSpace to upload in this manner is done using the gsUploadByGet function and passing it a configuration object that contains:
The url should just be an unencoded string. It will be URI encoded by the gsUploadByGet function before transmission. The optional filename will be the default filename for saving the file, and the user can override this within GenomeSpace. If a filename is not provided, it will use any part of the URL trailing the final '/' as the default filename. The optional successCallback and errorCallback functions will be called after the file upload has completed or had an error. Since these require JavaScript calling between browser windows, they should not be depended upon, but may be used to provide some additional feedback to the user. The successCallback and errorCallback functions will be passed as a single argument with the GenomeSpace directory path to which the file was saved (or where it failed to save, in the case of an error). In the example, the save via GET was performed by this function:
function uploadUrlByGetToGS(){
var dest = "http://beta.flymine.org/beta/service/query/results?format=tab&start=0&query=%3Cquery+model%3D%22genomic%22+view%3D%22Gene.symbol+Gene.organism.name+Gene.chromosomeLocation.locatedOn.primaryIdentifier+Gene.chromosomeLocation.start+Gene.chromosomeLocation.end+Gene.proteins.name+Gene.exons.primaryIdentifier%22+%3E%3Cconstraint+path%3D%22Gene.chromosomeLocation.locatedOn%22+type%3D%22Chromosome%22+%2F%3E%3Cconstraint+path%3D%22Gene.length%22+op%3D%22%26lt%3B%22+value%3D%228000%22+%2F%3E%3Cconstraint+path%3D%22Gene.pathways.name%22+op%3D%22ONE+OF%22+%3E%3Cvalue%3EMetabolic+pathways%3C%2Fvalue%3E%3Cvalue%3EGene+Expression%3C%2Fvalue%3E%3Cvalue%3ETranscription%3C%2Fvalue%3E%3Cvalue%3EmRNA+Processing%3C%2Fvalue%3E%3C%2Fconstraint%3E%3Cconstraint+path%3D%22Gene.chromosome.primaryIdentifier%22+op%3D%22%3D%22+value%3D%222L%22+%2F%3E%3C%2Fquery%3E&_=1362507.";
var onComplete = function(savePath){
alert('outer Saved to GenomeSpace as ' + savePath);
};
var onError = function(savePath){
alert('outer ERROR saving to GenomeSpace as ' + savePath);
};
gsUploadByGet({
url: dest,
filename: 'TheFileName.txt',
successCallback: onComplete,
errorCallback: onError
});
return false;
};
Sending a File via a POSTIn cases where there is not a public, unprotected URL that GenomeSpace can GET the file from, the server may instead POST the file to GenomeSpace so that the data source does not need to expose the file on its server. The POST may also be used where the data to be sent to GenomeSpace is dynamically generated and does not have a persistent URL from which it can be retrieved. In this method the return from the POST is a JSON object with one field, completionUrl, which is a URL that the POSTing server must open in a browser to allow the user to complete the sending of the file to GenomeSpace. What happens in this case is:
It is important to note here that after the file is POSTed and until it is saved by the user, it is only protected by the obscurity of its URL (a random number appended to the filename). If this level of security is not sufficient, you must NOT use this method but instead perform a complete GenomeSpace tool integration to ensure that all transfers are only done via HTTPS and are properly secured at all times. See the developer documentation for details (http://www.genomespace.org/adopters). When POSTing the file in the first step, the name of the file parameter is not significant. The receiving method will only accept one file per POST, and will always take the first file that it finds, no matter what the HTTP parameter name is. Thus, only the first file will be saved and made available for download. If you are POSTing the files to the GenomeSpace upload from JavaScript, your page will also need to include the JQuery JavaScript library. This is because the gsUploadByPost method is using the JQuery ajax function. The specific version of JQuery is probably not important due to the limited use, but the method has been tested with JQuery 1.7.2. Appendix: gsuploadwindow.jsThe gsuploadwindow.js script has the JavaScript methods for doing the POST or GET from JavaScript to GenomeSpace. Developers who want to POST to GenomeSpace from a server (i.e., not from JavaScript) can look at the details of the Ajax call as an example.
// get the path to this script at load time
// so we know what GenomeSpace UI to send the files to
var scripts= document.getElementsByTagName('script');
var path= scripts[scripts.length-1].src.split('?')[0]; // remove any ?query
var mydir= path.split('/').slice(0, -2).join('/')+'/'; // remove last filename part of path and the /uploads directory
var jsuiRoot = mydir;
function gsUploadByGet(config){
// expect an object with url, filename (optional), successCallback(optional), errorCallback (optional)
var gsUploadUrl = jsuiRoot +"/upload/loadUrlToGenomespace.html?uploadUrl=";
var dest = encodeURIComponent(config.url);
var filenameParam = "";
if (config['filename'] != null){
filenameParam = "&fileName="+config.filename;
}
var newWin = window.open(gsUploadUrl + dest + filenameParam, "GenomeSpace Upload", "height=340px,width=550px");
newWin.focus();
if (config['successCallback'] != null) newWin.setCallbackOnGSUploadComplete = config['successCallback'];
if (config['errorCallback'] != null) newWin.setCallbackOnGSUploadError = config['errorCallback'];
}
function gsUploadByPost(formData){
// expects an object with formData( the file(s)),
function gsUploadByPostErrorHandler(a,b,c,d){
alert('An error occurred psting the file to GenomeSpace');
};
function gsUploadByPostCompleteHandler(responseObj,status,xhr){
if (typeof responseObj == "string"){
responseObj = JSON.parse(responseObj);
}
var newWin = window.open(responseObj.completionUrl, "GenomeSpace Upload", "height=340px,width=550px");
newWin.focus();
};
$.ajax({
url: jsuiRoot + '/postToGenomeSpace', //server script to process data
type: 'POST',
crossDomain: true,
success: gsUploadByPostCompleteHandler,
error: gsUploadByPostErrorHandler,
data: formData,
//Options to tell JQuery not to process data or worry about content-type
cache: false,
contentType: false,
processData: false
});
}
|