Java Client Development Kit (CDK) (version beta 4.2)1 IntroductionThe public interface to GenomeSpace includes several web service providers: the Identity Manager, the Data Manager, and the Analysis Tool Manager. The GenomeSpace Client Development Kit (CDK) simplifies access to these services. The CDK provides an API for GenomeSpace clients to login, manage users, manage files, and launch web tools. Non-Java clients may use the same functionality via the REST web services API. The REST web services accessed by the CDK are described in documents found in the sections called “GenomeSpace Service Interfaces” in http://www.genomespace.org/adopters. For CDK usage examples, please refer to the JUnit tests that can be found in the Bitbucket version control system at https://bitbucket.org/GenomeSpace/combined. Look for Java source files in the directory combined/cdk/src/test/java/org/genomespace/client/test. 2 Requirements2.1 Identity ManagementThe CDK provides an API for GenomeSpace clients to:
2.1.1 LoginThe login method takes the username and password, sends those securely to the IdentityManager, receives the encrypted GenomeSpace user token from the IdentityManager, holds onto that token, and hides the token from the client. If the login fails, then the exception is returned to the client. 2.1.2 LogoutThe logout method clears the GenomeSpace user token. 2.1.3 Query Login StatusThe CDK provides the client with the ability to determine whether the current user is logged into GenomeSpace. 2.1.4 Register UserThe CDK provides the client with the ability to register a new GenomeSpace user. This method takes a username, password, and e-mail address, sends that information securely to the IdentityManager, and returns to the client whether the registration succeeded or not. If the registration fails (for example, if the requested username is already in use), then the CDK returns to the user an understandable error message. 2.2 File ManagementThe CDK provides the client with the ability upload files to GenomeSpace, download files from GenomeSpace, list files in GenomeSpace, and delete files in GenomeSpace. The CDK only provides access to files owned by the user – files owned by other users are not visible to the user. 2.2.1 UploadThe CDK provides the ability to upload a file to GenomeSpace. The upload method takes as input the location of the file to be uploaded, then uploads that file to the user’s GenomeSpace home directory. 2.2.2 DownloadThe CDK provides the ability to download a file from GenomeSpace. There are two forms of the download operation – one form actually downloads the file to a temporary directory and returns the location of that temporary file; the other form returns a signed, one-time usage URL that the client can use to directly download the file from Amazon S3. The download methods throw an appropriate exception if the file is not found on GenomeSpace. 2.2.3 File ConversionThe CDK provides the ability to download application data files in any of the different data formats that the Data Manager web service is able to transform the original file into. 2.2.4 ListThe CDK provides the ability to list the files in the user’s GenomeSpace home directory. This method returns a representation of the files in GenomeSpace, such that the objects can be easily passed to the download or delete method. 2.2.5 DeleteThe CDK provides the ability to delete files in the user’s GenomeSpace home directory. This method takes the name of the file as input and delete that file in GenomeSpace. If the file is not found on GenomeSpace, it throws an appropriate exception. 2.3 Analysis Tool ManagementThe CDK is responsible for providing GenomeSpace clients with web tool launch capabilities. The GenomeSpace Analysis Tool Manager (ATM) maintains a repository of tools available via the web that can be launched on behalf of a user. These tools may be web applications or client applications that run on the user’s local machine, but are launchable via an URL. 2.3.1 List Web ToolsThe CDK provides the client with the ability to retrieve a list of the available web tools available in the ATM. 2.3.2 Launch URL for a Web ToolThe CDK provides the client with the ability to retrieve an URL to launch a web tool, passing it GenomeSpace files as arguments. 3 Implementation3.1 DescriptionThe CDK provides a client application with the ability to perform basic identity management tasks and basic GenomeSpace file management tasks. The identity management tasks include the ability to register a new user, login, logout, and check whether the current user is logged in. The file management tasks allow the client to upload a file to GenomeSpace, download a file from GenomeSpace, list files uploaded to GenomeSpace, and delete a file uploaded to GenomeSpace. The main entry point to the CDK is the GsSession class. The client instantiates a GsSession instance, and then calls methods on GsSession to perform identity and file management operations. 3.2 GsSession ConstructionThe GsSession class provides several constructors. Most clients will simply use the default constructor, which configures the GsSession instance to communicate with the production GenomeSpace servers running on Amazon AWS. Prior to instantiating a session, you should identify your tool to GenomeSpace. This is optional but allows for more detailed tracking of tool use within the system. GsSession.setToolName("MyToolName");Several other constructors are provided for development purposes. These constructors provide the ability to specify the URL for GenomeSpace servers, so that the JCDK can be used for testing development servers. In addition, a constructor is provided that takes a GenomeSpace token as an argument, for use in testing GenomeSpace authentication. These other constructors are not needed for development of GenomeSpace clients and consequently are not specified in this document. 3.3 Identity OperationsThe GenomeSpace web services (Identity Manager, Analysis Tool Manager, etc.) require that the user be logged into GenomeSpace in order to access their services. 3.3.1 LoginTo login using the CDK, the client should create a GsSession object and then call the login method. The login method takes the username and password as arguments, encrypts them, and sends them to the IdentityManager for authentication. The login method returns a User object that contains the username: GsSession session = new GsSession();Assuming that authentication is successful, the IdentityManager then returns an authentication token in an HTTP cookie. GsSession includes the cookie value in every HTTP request to the GenomeSpace web services. The token contains the username and an expiration time stamp. If the token expires, the web services will return an HTTP authentication failure, and the CDK will throw an AuthorizationException. The client holds onto the GsSession object, because that is required for any further identity and file operations, as described below. To ensure a standard GenomeSpace look and feel in desktop applications (web applications using the CDK will presumably use OpenID) and for developer convenience, a GenomeSpace login dialog is provided in the CDK.
When using the GSLoginDialog, you should always first use the isLoggedIn method (see section 3.2.2 below) first to prevent showing the dialog when a cached reusable identity token already exists.
session = new GsSession();
if (session.isLoggedIn()){
// if we were logged in by another app on this machine we can get the cached username
lblUsername.setText(session.getCachedUsernameForSSO());
} else {
// open a GSLogin Dialog
final GSLoginDialog loginDialog = new GSLoginDialog(null, Dialog.ModalityType.APPLICATION_MODAL);
loginDialog.setVisible(true);
final String userName = loginDialog.getUsername();
final String password = loginDialog.getPassword();
try {
session.login(userName, password);
return;
} catch (final AuthorizationException e) {
JOptionPane.showMessageDialog(null, "Invalid user name or password!","Login Error",
JOptionPane.ERROR_MESSAGE);
}
}
More detail on the GSLoginDialog is available in section 4 of this document. 3.3.2 isLoggedInThe CDK provides the isLoggedIn method so that a client can determine whether their current GsSession is logged into GenomeSpace. This method returns true if the GsSession is logged into GenomeSpace and false otherwise: GsSession session = new GsSession();3.3.3 LogoutTo logout using the CDK, the client should call the logout method on their GsSession object: GsSession session = new GsSession();3.3.4 Register UserThe GsSession object provides the client with the ability to initiate registering a new user in GenomeSpace. After this call is done, an email is sent to the user (using the email address passed) which contains a URL to a confirmation page. This method requires a username, password, and e-mail address: GsSession session = new GsSession();3.3.5 Get All UsernamesThe client can get all GenomeSpace usernames. For security, a user login is required. It may be any user. GsSession session = new GsSession();3.3.6 Get User InformationThe client can get email and other details of a GenomeSpace user. For security, that user must be logged in. GsSession session = new GsSession();3.3.7 Change User PasswordThe client can change the password of a GenomeSpace user. For security, that user must be logged in. GsSession session = new GsSession();3.3.8 Change User EmailThe client can change the password of a GenomeSpace user. For security, that user must be logged in. GsSession session = new GsSession();3.3.9 Get Remaining Token Time
The client can find out the number of milliseconds until the user must login again due to an expired token. GsSession session = new GsSession();This version takes an explicit token: GsSession session = new GsSession();3.3.10 Get Username from TokenThe client can find out the username from a token: GsSession session = new GsSession();3.3.11 Reset User PasswordThe client can cause a temporary password to be assigned to a user and emailed to their email address on record. The temporary password expires after one day or after the first successful login. This operation does not affect the normal password, which may be changed when user logs in with the temporary. Does not require a user to be logged in. GsSession session = new GsSession();3.3.12 Remove UserThe client can remove a GenomeSpace user. For security, that user must be logged in. GsSession session = new GsSession();3.4 Client Defined UserProfilesGenomeSpace provides persistent storage for data that a client may want to use in their application which is associated with a given user. This is data for the client’s use, and is not used by GenomeSpace for any purpose. Internal to GenomeSpace this data put in a UserProfile object, and the data must be presented as a String datatype. Currently the size limit for one UserProfile is about 250KB, depending on the number of optional attributes put on the UserProfile (each one reduces size by 1KB). The CDK has an API for storing and retrieving lists of strings in a UserProfile, and also an API for the raw UserProfile. The difference is in the number of identifiers that the client wants to use. The List API has two levels of identity, a “type” and an “identifier”, and has a fetch method for getting all UserProfiles of a given type, and by specific identifier. The raw API simply exposes a Map of optionalAttributes that must be managed by the client. 3.4.1 UserProfile List APIThe client can persist an arbitrary list of strings for a user in one UserProfile. Multiple UserProfiles are allowed per user. For security, the user associated with the UserProfile must be logged in. 3.4.1.1 Create UserProfile ListThe client can create a UserProfile for storing a List of String. For security, the user associated with the UserProfile must be logged in. // Define the client-defined identifiers3.4.1.2 Retrieve All UserProfile List having TypeThe client can retrieve all UserProfile lists for a user having the specified listType. For security, the user associated with the UserProfile must be logged in. String namespace = this.getClass().getName();3.4.1.3 Retrieve One UserProfile ListThe client can retrieve one specific UserProfile list by giving both the type and the identifier. For security, the user associated with the UserProfile must be logged in. String namespace = this.getClass().getName();3.4.1.4 Delete UserProfile ListThe client can delete a UserProfile. For security, the user associated with the UserProfile must be logged in. String namespace = this.getClass().getName();3.4.2 UserProfile Raw APIThe client can persist an arbitrary string for a user in one UserProfile, with arbitrary optional attributes used to identify the UserProfile. Multiple UserProfiles are allowed per user. For security, the user associated with the UserProfile must be logged in. 3.4.2.1 Create UserProfileThe client can create a UserProfile for storing a String. For security, the user associated with the UserProfile must be logged in. String persistedValue = "some string, possibly a marshalled or serialized object";3.4.2.2 Retrieve UserProfileThe client can retrieve UserProfiles for a user matching the specified optionalAttributes. For security, the user associated with the UserProfile must be logged in. Map<String, String> optionalAttributes = new HashMap<String, String>();3.4.2.3 Retrieve One UserProfileThe client can retrieve one specific UserProfile list by giving the profileId that was generated when the UserProfile was created. For security, the user associated with the UserProfile must be logged in. GsSession session = new GsSession();3.4.2.4 Delete UserProfileThe client can delete a UserProfile by profileId, which is needed for uniqueness. For security, the user associated with the UserProfile must be logged in. GsSession session = new GsSession();3.5 File Access Group ManagementGenomeSpace filesystem has access restricted by user and by group. A group is a collection of users. Use this API to create, lookup, update, and delete groups and group membership. Applying access permissions to a group is handled by the DataManager and is described elsewhere. 3.5.1 Create GroupThe client can create a group of GenomeSpace users. Requires a user to be logged in. If no explicit members are given, then the logged-in user becomes the first member and also the group’s administrator. If members are given then those become the members. There must be at least one member and at least one administrator for every group; the group will not be created otherwise. GsSession session = new GsSession();3.5.2 Find Group by IdThe client can lookup a group of GenomeSpace users. Requires a user to be logged in. Any user will do, but group membership will be shown only if the user is a member of the group, or if the group is non-member readable. GsSession session = new GsSession();3.5.3 Find Group by List of IdThe client can lookup a group of GenomeSpace users. Requires a user to be logged in. Any user will do, but group membership will be shown only if the user is a member of the group, or if the group is non-member readable. Returns the groupId and the Group. GsSession session = new GsSession();3.5.4 Find GroupIds for UserThe client can lookup groupIds of all groups that the user is a member of. Requires a user to be logged in. Any user will do. GsSession session = new GsSession();3.5.5 Find All Group Names and IdsThe client can lookup all of the groups. Requires a user to be logged in. Any user will do. GsSession session = new GsSession();3.5.6 Delete GroupThe client can delete a group. Requires a group administrator to be logged in. GsSession session = new GsSession();3.5.7 Update GroupThe client can update a group. Requires a group administrator to be logged in. GsSession session = new GsSession();3.5.8 Add Group MemberThe client can add a member to an existing group. Requires a group administrator to be logged in, or a group member to be logged in AND the group to be self-administered. GsSession session = new GsSession();3.5.9 Delete Group MemberThe client can delete a member from an existing group. Requires a group administrator to be logged in, or a group member to be logged in AND the group to be self-administered. GsSession session = new GsSession();3.6 File OperationsGenomeSpace provides each user with a private base directory in S3; the users only have access to their own GenomeSpace files. The CDK allows users to:
All the file system operation are defined by the interface: 3.6.1 Default DirectoryEvery user has a personal directory assigned to them in the GenomeSpace file system. In this directory and all directories below it, the user is permitted all the supported file operations. The default file path to the personal directory is /Home/username (but you should depend on the API instead of /Home/username in your code). To obtain a listing of the user’s personal directory: DataManagerClient dmClient = gsSession.getDataManagerClient();
The initial default directory for users is currently different from the personal directory. It includes the personal directory as well as directories shared with everyone and with the specific user. The default directory is /Home (but you should depend on the API instead of /Home in your code).
DataManagerClient dmClient = gsSession.getDataManagerClient();The GSDirectoryListing is discussed below. 3.6.2 Listing Directory ContentsFor a given directory file path: String thePath = “/users/test”;GSDirectoryListing has a reference to the metadata (GSFileMetadata) of the target directory and a list of metadata objects corresponding to each child file and directory. GSFileMetadata indicates whether a particular file system object is a file or directory, when it was last modified, the size in bytes, etc. It also carries format information, including the format of the file (if Data Manager is able to recognize it) and the different data formats to which this file can be converted. If you have the metadata object for the directory, the preferred way of getting a directory listing is:
GSDirectoryListing dirContents = dmClient.list(dirMetadata);
3.6.3 Obtaining Metadata for a File System Object by its PathThe file system metadata objects can be obtained indirectly through directory listings or at object creation time. They can also be obtained through their paths. String objectFilePath = “/users/test/myFileOrDirName”;3.6.4 Creating DirectoriesThe CDK allows arbitrary directory levels. To create a directory, in this case, as the child of the home directory: GSFileMetadata homeDirMeta = dmClient.listDefaultDirectory().getDirectory();3.6.5 Uploading FilesThe CDK supports two methods for this operation. The simplest, if you have the GenomeSpace target directory metadata and you want to upload a local file, preserves the same name remotely: GSFileMetadata newDirMeta = dmClient.createDirectory(homeDirMeta,newDirName);If you want to assign a different name to the uploaded remote file: GSFileMetadata newDirMeta = dmClient.createDirectory(homeDirMeta,newDirName);3.6.6 Downloading FilesTo download a file, you need to get the metadata object of the source remote file: GSDirectoryListing dirListing = dmClient.listDefaultDirectory();The Boolean argument overwriteIfExists will come in to play if the target file exists. An exception will be raised if flag is false and the target file already exists in the local file system. 3.6.7 Downloading Files in a Specific Data Format/Perform a Conversion on a Remote FileThe Data Manager is able to perform format conversions on files. You can request a download in any of the formats listed in the GSFileMetadata object. GSDirectoryListing dirListing = dmClient.listDefaultDirectory();To download the file in a particular format, you will need to specify the format and the destination file or the destination directory. GSDirectoryListing dirListing = dmClient.listDefaultDirectory();
You can also call the method
In this case, the local file name will be determined by the Data Manager server. You just specify to what directory you want the file saved.
Finally, if you just want a
or
For all the methods discussed above, leaving 3.6.8 Obtaining a URL to Download Original or Converted FilesYou can obtain a URL that can be used by your application to access the original file or converted file directly, however your application will have will have to be able to deal with:
You can use one of three methods:
The URLs for the remote files and for the data formats are available by calling getUrl() on GSFileMetadata and GSDataFormat. 3.6.9 Deleting Files and DirectoriesIf the user owns the file or directory (i.e., the file or directory is under her home directory), delete files/directories using: GSDirectoryListing dirListing = dmClient.listDefaultDirectory();The procedure is the same for files and directories. Only empty directories can be deleted. 3.6.10 Copying Files and DirectoriesFiles and directories can be copied within the GenomeSpace file system without having to download them. GSDirectoryListing dirList = dmClient.listDefaultDirectory();In the case of the directories, the procedure is the same. The Data Manager will recursively copy the directory to the source directory under the target parent directory with the newly assigned file name. A null for new file name will keep the original file object name. 3.6.11 Moving Files and DirectoriesSame as the copy, but the source objects will be removed. As before, directory moves are recursive. GSDirectoryListing dirList = dmClient.listDefaultDirectory();3.7 File Permissions and Ownership
Ownership of files and directories is established by who is the owner of the top level directory. Any file and directory that is directly or indirectly under a user's home directory is owned by that user. As such, the owner user is able to read, write, delete, and grant permissions on any object below. Data Manager supports sharing of files and directories through the use of access control lists (ACLs). The owner of a file or directory can grant read and write permissions to any other user or group. Each ACL points to the object with which it is associated and includes a list of access control entries (ACE) identifying the user or group (generically referred as a Security Identity [SID]) and the permission that has been granted. Read permission on a file means that its contents can be downloaded. Write permissions mean that the file can be either updated/replaced or deleted. On a directory, read permission mean that the grantee can list the contents of the directory. Write permission mean that they can upload new files and delete and update existing ones.
ACL grants are inherited down the directory structure. So if you grant read permissions on a parent directory to another user, that user will be able read every file and directory below it in the hierarchy. 3.7.1 Granting PermissionsTo grant permissions, the logged-in user needs to be the owner of the file (i.e., the file needs to be under the granter’s home directory). //You will need to get a hold of the file or directory GSFileMetadata object.3.7.2 Listing and Checking Permissions on a File or DirectoryIf you just want to show the effective permissions on a file or directory, the easiest thing to do is get the effectiveAcl property from the file object GSFileMetadata. Keep in mind that this is not a “real” ACL object, but rather a synthetic ACL that captures permissions granted on the file directory, and that have been inherited from the directory structure and reflects ownership of the object. You cannot delete this ACL object or revoke the permissions listed here, but you will be able to know if you can list a directory, delete a file,etc. //You will need to get a hold of the file or directory GSFileMetadata object.//We have added a helper class to build SecurityIdentityTo get the “real” ACL objects, such as in the case where you want to enable editing permissions, you can use: Acl theFileAcl = dmClient.getAcl(gsFileMetatadata); Perhaps even more useful is getting all the ACLs associated with a file object, both directly connected and inherited: List<Acl> acls = dmClient.getAclHierarchy(gsFileMetadata); Each ACL has a method called getObjectId(), which returns the full file path of the file object that it is connected to. 3.7.3 Revoking PermissionsYou can revoke a permission that has been previously granted. Be aware though, that if a permission has been granted to a directory further up in the hierarchy, you cannot take away that permission from the object below. For example, if a directory is granted write permissions, you will not be able to revoke write permissions on a single file in the directory. Two ways to revoke, the first is similar to one of the grant methods presented above. dmClient.revoke(gsFileMeta,securityIdentity,Permission.READ,Permission.WRITE); To reiterate, this will only work for permissions that are directly attached to the file or directory, not for inherited permissions. If you have the ACL object on hand, obtained via either the getAcl() or getAclHierarchy() calls, then you can get a hold of the access control entries. You can revoke a permission by deleting the associated access control entry (ACE). Here is a snippet that removes all permissions granted to a user on a file: Acl theAcl = dmClient.getAcl(gsFileMeta);3.8 Analysis Tool OperationsThe GenomeSpace CDK provides client the ability to:
Later versions of the CDK will add additional functionality. The client must be logged into the CDK in order to use the Analysis Tool operations. 3.8.1 List Web ToolsThe client can retrieve a list of web tools using the AnalysisToolManagerClient, which can be obtained from the GsSession object. The getWebTools method returns a list of WebToolDescriptors. AnalysisToolManagerClient client = gsSession.getAnalysisToolManagerClient();3.8.2 Launch URL for a WebToolThe CDK provides the client with the ability to retrieve a URL to launch a web tool, passing it GenomeSpace files as arguments. Once the client has retrieved a list of web tools, the client can select the webtool of interest and obtain a launch URL for that web tool using the getWebToolLaunchUrl method. DataManagerClient dmClient = gsSession.getDataManagerClient();3.8.3 Get WebToolThe CDK provides the client with the ability to retrieve a WebToolDescriptor by name. AnalysisToolManagerClient client = gsSession.getAnalysisToolManagerClient();
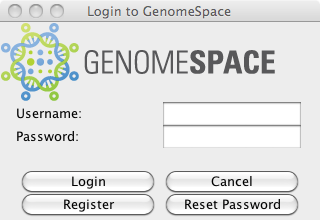
4. User Interface Elements of the CDKSome common user interface (UI) dialogs have been incorporated into the CDK for use by desktop applications. These have been implemented using Java Swing and include a login dialog and a file browser suitable for selecting a file for download and for selecting a parent file path to be used to upload a new file to a particular directory. These UI elements are in the CDK jar file in the java package org.genomespace.client.ui 4.1 GSLoginDialog
When using the GSLoginDialog, you should always use the isLoggedIn() method (see section 3.2.2) first to prevent showing the dialog when a cached reusable identity token already exists.
session = new GsSession();
if (session.isLoggedIn()){
// if we were logged in by another app on this machine we can get the cached username
lblUsername.setText(session.getCachedUsernameForSSO());
} else {
// open a GSLogin Dialog
final GSLoginDialog loginDialog = new GSLoginDialog(null, Dialog.ModalityType.APPLICATION_MODAL);
loginDialog.setVisible(true);
final String userName = loginDialog.getUsername();
final String password = loginDialog.getPassword();
try {
session.login(userName, password);
return;
} catch (final AuthorizationException e) {
JOptionPane.showMessageDialog(null, "Invalid user name or password!","Login Error",
JOptionPane.ERROR_MESSAGE);
}
}
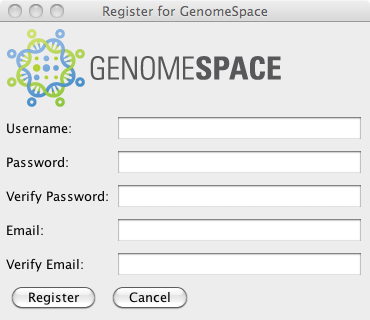
In the example above, we use a GsSession seperate from the login dialog. It is also possible to get a GsSession back directly from the login dialog: GSLoginDialog loginDialog = new GSLoginDialog(null, Dialog.ModalityType.APPLICATION_MODAL); loginDialog.setVisible(true); GsSession session = loginDialog.getSession(); The GSLoginDialog also includes a button to launch the GsRegistrationDialog.
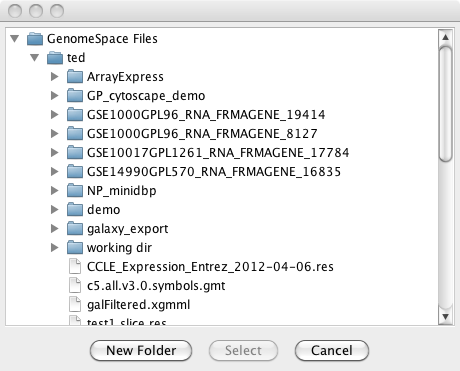
4.2 GSFileBrowserDialog
The GSFileBrowserDialog is a subclass of JDialog that allows a desktop application to browse through a tree representation of a logged-in user's GenomeSpace file system in the DataManager. GSFileBrowserDialog fileBrowser = new GSFileBrowserDialog(null, session.getDataManagerClient(), GSFileBrowserDialog.DialogType.FILE_SELECTION_DIALOG); fileBrowser.show(); GSFileMetadata selection = fileBrowser.getSelectedFileMetadata(); // Do something with the selected file The GSFileBrowserDialog can be opened in one of three dialog types. It can be a selection dialog, a save dialog, or a deletion dialog. It is up to the application to perform the actual actions upon a file selection while the dialog types simply change the title of the dialog. |