New data source: the cBioPortal for cancer genomicsGenomeSpace is proud to announce the addition of another new data source to the GenomeSpace tool catalog: the cBioPortal for cancer genomics from the Memorial Sloan Kettering Cancer Center. See the Loading Data from cBioPortal to GenomeSpace recipe to learn how to send your cBio query results to your GenomeSpace account. The cBioPortal for Cancer Genomics provides visualization, analysis and download of large-scale cancer genomics data sets such as The Cancer Genome Atlas (TCGA).
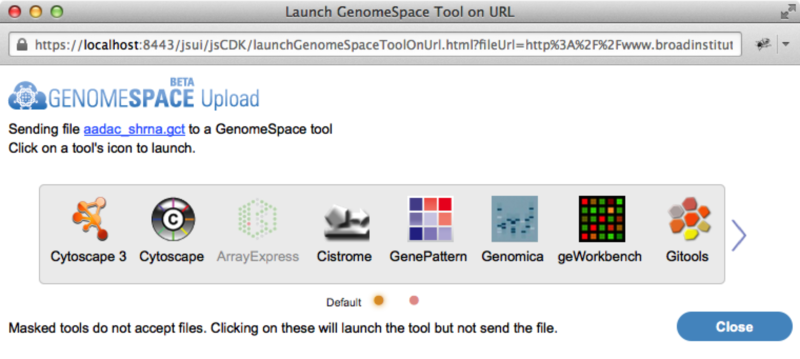
The GenomeSpace Web Development KitTool Developers: We are glad to announce the release of our GenomeSpace Web Development Kit (WDK), which allows web applications to easily save files to GenomeSpace. Applications can either provide URLs or send file contents to GenomeSpace in a POST, to be saved as a file. The WDK supports launching a GenomeSpace tool on a file or URL, so with minimal modification any web application can send data into the universe of GenomeSpace tools for analysis. Click here to see code samples, documentation, and a live demo to add this functionality to a website. Launching a GenomeSpace toolWhen users launch a WDK on a URL, they will see the following window. GenomeSpace authentication is handled in this window, obviating the need for web applications to integrate into GenomeSpace single sign-on.
Selecting a tool in the toolbar will launch the tool allowing the user to analyze the file. Please note that the current implementation does not yet support automatic format conversion. This will be added shortly. For more use cases and example code, see the live demo page.
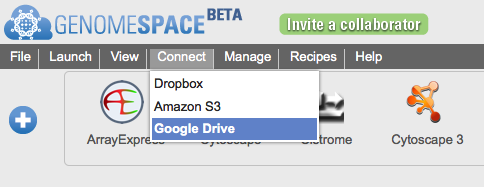
Connecting GenomeSpace to Google DriveThe GenomeSpace team is pleased to announce that you can now connect your GenomeSpace account to your Google Drive account. This gives you an additional option for accessing and analyzing files you have stored in the cloud. Instructions to connect are detailed in screenshots below. Briefly, to connect your GenomeSpace and Google Drive accounts:
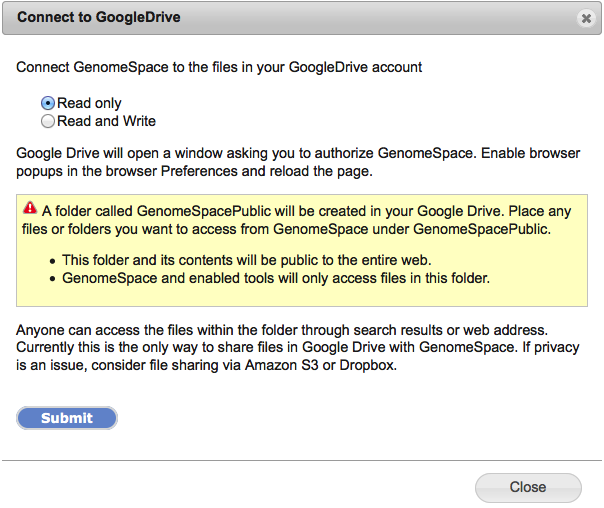
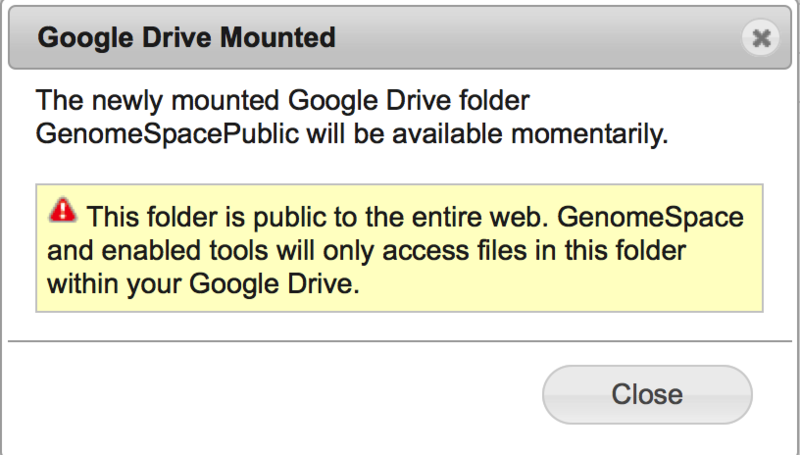
Approving access creates a new folder in Google Drive named "GenomeSpacePublic". GenomeSpace will operate exclusively on files and folders within GenomeSpacePublic, ignoring everything else in Google Drive. Place any data files you want to access from GenomeSpace tools under this folder. Please be aware the GenomeSpacePublic folder is created with "Public on the web" Google Drive permissions. This permission level allows anyone to access the contents of the folder via search results or the web address. If you have sensitive data that you would like to use from GenomeSpace data tools, we recommend you place it under your GenomeSpace home directory, in a GenomeSpace-mounted Dropbox account, or in an Amazon S3 bucket. Detailed instructions to connect GenomeSpace with Google DriveFirst, select Connect>Google Drive in the menu bar.
Next, select the privileges GenomeSpace can have in Google Drive. This can be Read only, but choose Read and Write if you want GenomeSpace to be able to send files into your Google Drive folders.
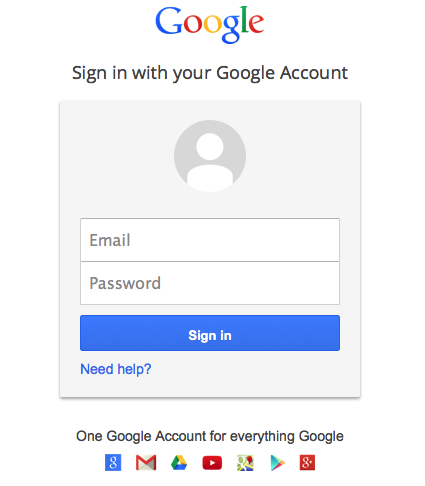
When you click Submit in this dialog, a new window will open to the Google Drive site for the next step. Note that if you have a popup blocker enabled on your browser, you have to allow this popup window to be opened. If you are not already logged in to Google, the Google window will ask you to login.
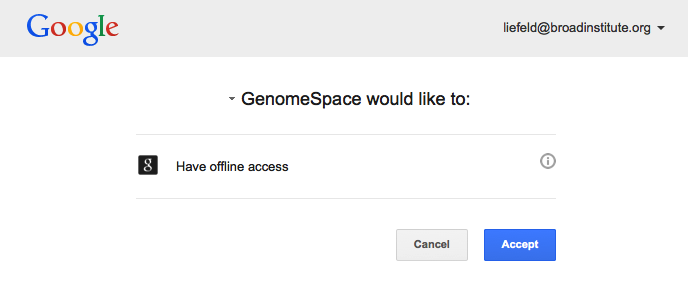
Google then asks you to confirm that you want to allow GenomeSpace access your Google account. Click Accept.
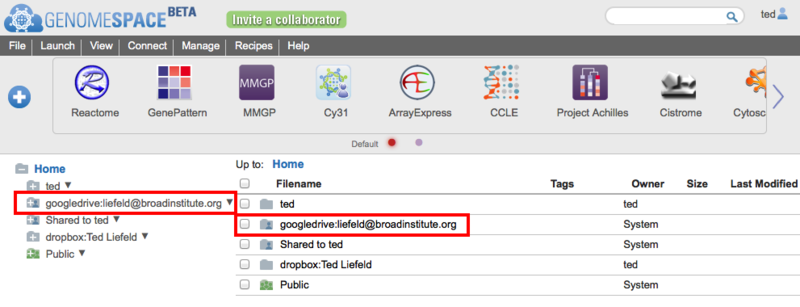
When you allow GenomeSpace access to your Google Drive account, GenomeSpace will connect to Google Drive and create a new folder in your GenomeSpace home directory.
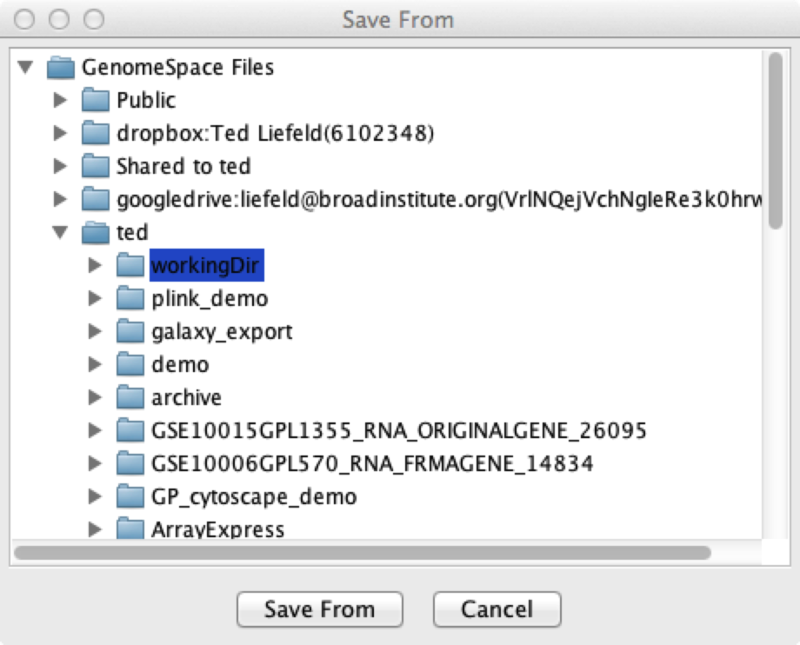
After this, you can copy files between GenomeSpace and Google Drive and send files directly from Google Drive to any of the GenomeSpace tools. If you want to disconnect Google Drive from GenomeSpace, right-click on the Google Drive folder and select Disconnect. For information on how to connect to your Dropbox account, refer to the Adding Dropbox page of the User Guide. For information on how to connect to Amazon S3 buckets, refer to the Adding an Amazon S3 Bucket page of the User Guide. Recursively downloading files from GenomeSpaceRecently a GenomeSpace user asked us how to recursively download all of the files in a directory. In this case, recursively means that they wanted to get everything from a folder, its subfolders, and their subfolders all the way down. The GenomeSpace UI itself will allow you to download every file in a directory, but it does not go into the subfolders within a directory - you still have to go to each of them one at a time. To make this easier we created an application you can use to log in to GenomeSpace, select a top level GenomeSpace directory where you want to start the download, and it will walk down the directory tree, copying the entire file and directory structure to your computer. You can download this application from the SharedData/Utilities/GSRecursiveDownloader folder on GenomeSpace. To use it, follow these steps;
Thats it!
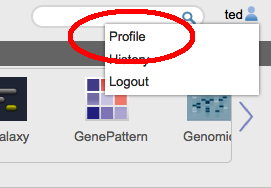
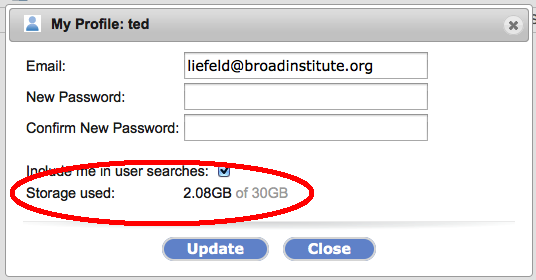
New Tool: ReactomeGenomeSpace is proud to announce the addition of another new tool to the GenomeSpace tool bar: Reactome, from the Ontario Institue for Cancer Research (OICR). REACTOME is an open-source, open access, manually curated and peer-reviewed pathway database. Pathway annotations are authored by expert biologists, in collaboration with Reactome editorial staff and cross-referenced to many bioinformatics databases. These include NCBI Gene, Ensembl and UniProt databases, the UCSC and HapMap Genome Browsers, the KEGG Compound and ChEBI small molecule databases, PubMed, and Gene Ontology. GenomeSpace StorageThe GenomeSpace team is happy to announce that we have increased the storage quota for all GenomeSpace users to 30GB. You can now also check how much storage you are using on GenomeSpace on the profile dialog. You can get to this from the 'profile' menu item near your username on the top right corner of the screen.
The profile dialog will display how much storage you are using:
If you need more than 30GB to store the data you want accessible to the GenomeSpace tools, you can also keep data files in your Dropbox or on Amazon S3 buckets that you own. If you need to exceed your quota for only a short term, you can contact us to request a temporary exception as well.
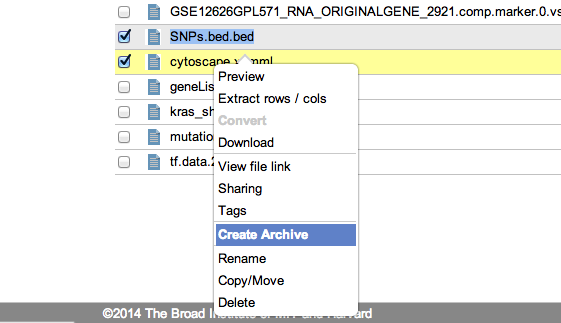
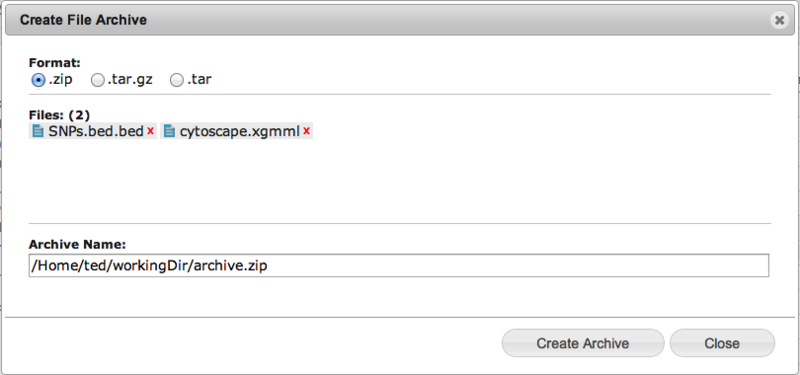
GenomeSpace now supports compression and decompression of your files in the cloudIn response to user requests, the GenomeSpace development team has added the ability to compress and decompress files and folders that are on your GenomeSpace storage. We are initially supporting .zip, .tar, and .tar.gz formats. Creating an archiveTo create an archive means to compress one or more files and folders. To start, simply select one or more files and folders in the GenomeSpace User Interface. You will see the new menu item, 'Create Archive' on both the popup (right click) and on the main File menus. If you include a folder to be compressed, the created archive will include all the contents of that folder. When the archive is created it copies the files into that archive, so your originals will be safe and unmodified by the archive creation.
Selecting either of these menu items will open up the 'Create File Archive' dialog box. Here you can select the type of compression to be used; .zip, .tar or .tar.gz and you can also choose the directory and filename for the new archive file.
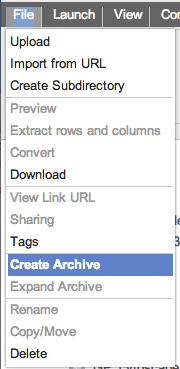
As the archive creation is taking place, a notification message Expanding an archiveExpanding (decompressing) an archive also begins from the file and/or popup menus. To start select the archive that you wish to decompress and then select the 'Expand Archive' menu as seen here. This menu item only shows up for files with the .zip, .tar and .tar.gz file extensions.
As with the compression, this will create a notifier at the bottom of the page to show you the status of the expansion That's it, please let us know if there are other features you would like to see added to GenomeSpace.
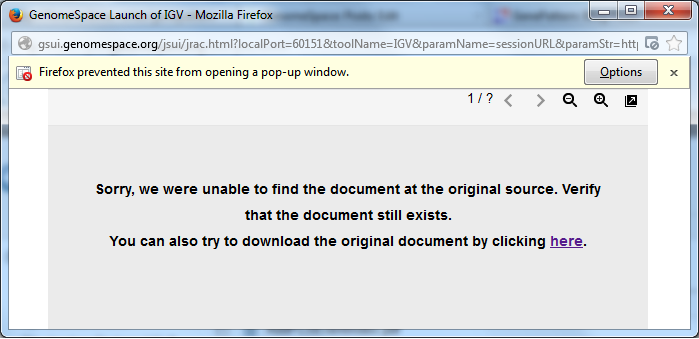
Unable to launch Desktop ToolsWe have recently heard from users that, with the recent Java and browser updates, GenomeSpace desktop tools (such as Genomica and IGV) are being blocked from running in browsers. You may have seen a window such as the one below after attempting to launch one of these tools: (The image below is from Firefox 27.0.1 running on Windows 7 64-bit)
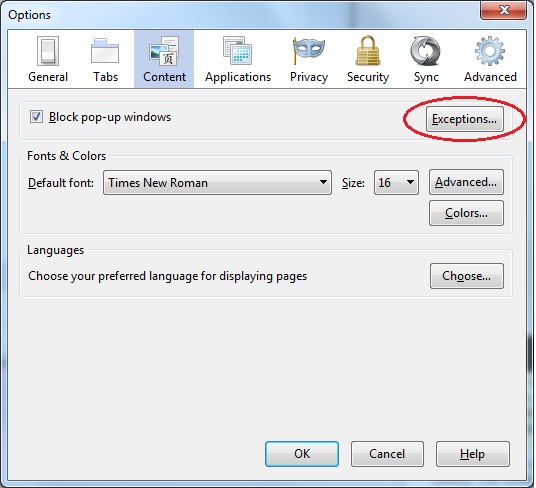
To resolve this issue, in any browser, you will need to turn off the pop-up blocker for GenomeSpace. For Firefox: On Windows or Mac, if you get the window you see above, click Options (Windows) or Preferences (Mac) and "Allow pop-ups from gsui.genomespace.org". You will then need to close that window and launch the tool again. From there the behavior should be as expected. If you did not get that window or it disappears before you can click on the Options or Preferences button:
For Chrome: You will likely only see the error window for a short period of time and will then need to manually add "gsui.genomespace.org" to the exception list, following the instructions here For Safari: As with Chrome, you will likely only see the error window for a short period of time. If pop-ups are blocked, nothing else will happen. If pop-ups are not blocked, you should see another browser window open and the jnlp file will be downloaded, per usual. To disable pop-up blocking in Safari:
Note that even after you have done this it may appear that nothing has changed; The error window may appear, a second browser window may be opened, but it may seem that nothing was downloaded. To check this, click on the download icon in the upper left of the Safari browser window.
You should see the launch file for the tool you have selected.
Please email us if these tips do not serve to resolve any issues you are having with the launching of GenomeSpace desktop tools. gs-help (at) broadinstitute.org
File Upload changesRecently a few members of the GenomeSpace community noticed that there was no way to know if a file upload was working or not after they had dragegd and dropped a file on a GenomeSpace folder. Turns out they had found a bug in the system. While an upload indicator was appearning for really large files, the notification system for smaller files was ... broken. We have now fixed the file upload notifications so that you can see indication of an upload in progress no matter what the size of the file (or files).
The upload indicator can be seen near the top right corner of your screen. If an upload is in progress it will be animated like this
We are working on making upload progress more visible and you should see more improvements in this part of the GenomeSpace UI shortly. GenomeSpace server and support coverage December 23 to January 2From December 21 to January 1, the GenomeSpace team will be unavailable due to the year-end closing of the Broad Institute. During this time, the GenomeSpace server will remain up and fully functional. We will be monitoring this daily and, should anything happen to disrupt service, we will work to resolve any issues within 24 hrs. However, due to the holiday closing, please be aware that:
If you have any questions, please feel free to contact us. |

 will appear at the bottom of the GenomeSpace User Interface (GSUI) showing the progress. Once it is complete, the notification will turn grey
will appear at the bottom of the GenomeSpace User Interface (GSUI) showing the progress. Once it is complete, the notification will turn grey  and you can click on it to have the GSUI navigate directly to the new archive. Should an error occur, the notification will turn red
and you can click on it to have the GSUI navigate directly to the new archive. Should an error occur, the notification will turn red 
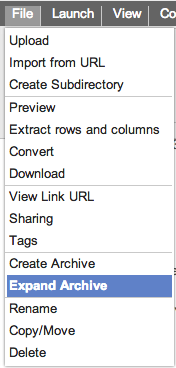
 which will turn grey when complete
which will turn grey when complete  . Clicking on the notification after it has completed will navigate the GSUI to the newly created file or folders that have been extracted from the archive.
. Clicking on the notification after it has completed will navigate the GSUI to the newly created file or folders that have been extracted from the archive.