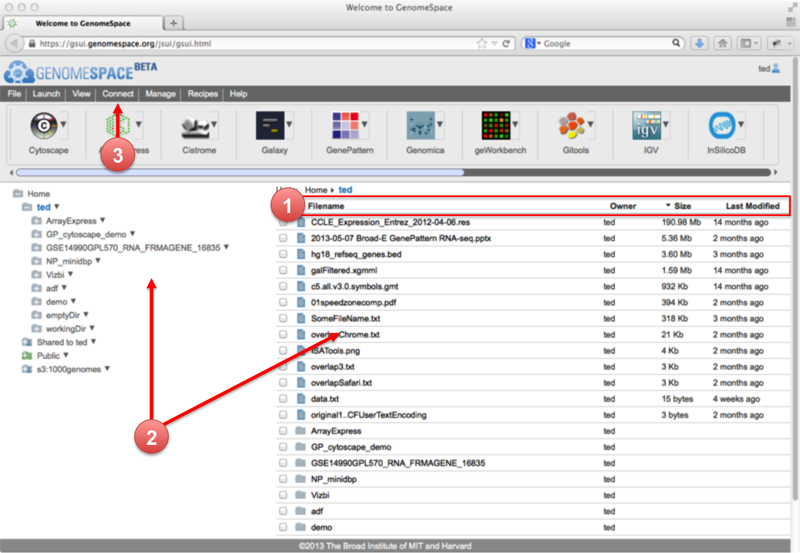
GenomeSpace UI enhancementsThis morning, we updated the GenomeSpace user interface (GSUI) with several enhancements to make it easier to use and work with your files. The new GSUI looks like this:
Some of the changes and upgrades from the previous version include the following:
If you have suggestions for additional improvements, please contact us at gs-help@broadinstitute.org. Uploading FilesThere are currently two methods for uploading files in GenomeSpace:
or
In general, we recommend dragging a file from your computer and dropping it on a GenomeSpace folder. However, if you are uploading files larger than 1 GB, you should use the Java Uploader. There are some known issues to be aware of:
Workaround: You can either try a newer browser or use the Java Uploader.
Workaround: For very large files, use the Java Uploader.
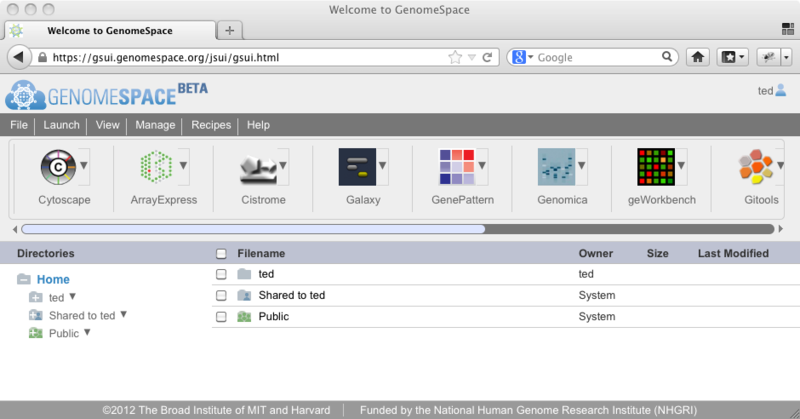
Workaround: Use Firefox. New Features: File manipulation enhancementsNew Feature: Drag and drop files to copy themSince GenomeSpace was released, you have been able to move a file or folder by using the mouse to drag and drop it into the folder you would like to move it to. As of the latest update, you can now use this same approach to copy a file (or folder) to a new location. To copy the file/folder instead of moving it, press and hold down the CTRL, SHIFT, or ALT key on your keyboard (any one key will do) while you drop the file into its new location. In addition, if you drag and drop a file or folder that you have read, but not write, access to, it will now copy the folder to the new location by default. We hope that these changes will make it even easier for you to manage your GenomeSpace files and folders. Please feel free to contact us at gs-help at broadinstitute dot org if you would like to request any other changes or features. Experimental Feature: Drag files from your desktop to uploadIn an effort to make it even easier to upload your data to GenomeSpace, we have enabled HTML5 uploading in GenomeSpace. What this means is that if you are using a modern browser that supports HTML5 (for example, Firefox 18 or higher, Chrome 24 or higher), you can now upload files to GenomeSpace by dragging them from your desktop and dropping them onto the folder in GenomeSpace where you would like them to be. Please note that this is still an experimental feature, so there may be some rough edges. If you happen to experience isssues, please let us know at gs-help at broadinstitute dot org and we will try to smooth them out for you. One warning for users of the Chrome browser: Chrome loads your files into memory during the upload process, so if you use this feature for very large files (e.g., 1GB+) the memory used by your Chrome process can become very large. We suggest that for very large files, you continue to use the Java Uploader applet at this time. New Tool: GitoolsGenomeSpace is proud to announce the addition of another new tool to the GenomeSpace tool bar: Gitools, from Biomedical Genomics Group at the Biomedical Research Park in Barcelona (PRBB). Gitools is an open-source tool that performs analyses and allows users to visualize data and results as interactive heatmaps that facilitate the integration of novel data with previous knowledge. Gitools can import data import from GenomeSpace, IntOGen, Biomart, Gene Ontology, and KEGG . GenomeSpace UI UpdatedThe GenomeSpace User Interface (GSUI) has received some minor updates and one bug fix this morning. To make the UI more in line with standard web applications, we have moved the menu bar to the top of the page above the toolbar. In addition, we have fixed a bug where the GSUI failed to properly re-open a directory after it has been renamed. The new GSUI now looks like this:
New Feature: Mounting Amazon AWS S3 BucketsThe GenomeSpace Data Manager was originally built to save the files you upload to GenomeSpace in an Amazon Simple Storage System (S3) bucket that is managed by GenomeSpace itself. However you can add additional Amazon S3 buckets to GenomeSpace that you or a third party has set up to make the file contents available to your GenomeSpace and your GenomeSpace tools. For buckets that are publicly accessible, you only need to tell GenomeSpace the name of the bucket to mount it. However, for private buckets, or those with limited non-public accessibility, the process is more complex, requiring you to set up a sub-account and the minimal permissions in Amazon to share the bucket with GenomeSpace. Once a bucket has been mounted in GenomeSpace, you can share it with other GenomeSpace users using the standard GenomeSpace sharing dialogs. For details on how to mount a bucket into your GenomeSpace, follow the steps in the documentation. Having Trouble With Dialog Boxes in GenomeSpace?Do you use the AdBlock Plus plugin for Firefox or Chrome? If so, you may have noticed some issues with empty or mispositioned dialog boxes in GenomeSpace. The solution: Select Tools>AdBlock Plus>Disable on gsui.genomespace.org, then restart your browser. You should start seeing the correct dialog boxes and functionality. Public Data: TCGA Ovarian CancerWe are pleased to announce that data from the following paper have been made available on GenomeSpace: The Cancer Genome Atlas Network. Integrated genomic analyses of ovarian carcinoma. Nature. 474 (7353):609-615. The GenomeSpace mirror of this dataset includes all public level 3 and level 4 data from https://tcga-data.nci.nih.gov/docs/publications/ov_2011/. You can access the data in GenomeSpace at the path /Public/SharedData/Datasets/TCGA/Ovarian Cancer/. (This requires a GenomeSpace login. If you don't have one, it's easy to register.)
Abstract Public Data: TCGA Colon and Rectal CancerWe are pleased to announce that data from the following paper have been made available on GenomeSpace: The Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature. 2012;487:330-337. [doi:10.1038/nature11252] The GenomeSpace mirror of this dataset includes all public level 3 and level 4 data from https://tcga-data.nci.nih.gov/docs/publications/coadread_2012/. You can access the data in GenomeSpace at the path /Public/SharedData/Datasets/TCGA/Colon and Rectal/. (This requires a GenomeSpace login. If you don't have one, it's easy to register.) Abstract:To characterize somatic alterations in colorectal carcinoma (CRC), we conducted genome-scale analysis of 276 samples, analyzing exome sequence, DNA copy number, promoter methylation, mRNA and microRNA expression. A subset (97) underwent low-depth-of-coverage whole-genome sequencing. 16% of CRC have hypermutation, three quarters of which have the expected high microsatellite instability (MSI), usually with hypermethylation and MLH1 silencing, but one quarter has somatic mismatch repair gene mutations. Excluding hypermutated cancers, colon and rectum cancers have remarkably similar patterns of genomic alteration. Twenty-four genes are significantly mutated. In addition to the expected APC, TP53, SMAD4, PIK3CA and KRAS mutations, we found frequent mutations in ARID1A, SOX9, and FAM123B/WTX. Recurrent copy number alterations include potentially drug-targetable amplifications of ERBB2 and newly discovered amplification of IGF2. Recurrent chromosomal translocations include fusion of NAV2 and WNT pathway member TCF7L1. Integrative analyses suggest new markers for aggressive CRC and important role for MYC-directed transcriptional activation and repression. Calling the GenomeSpace API from PythonFor today's technology tidbit, we're going to explore how to call the GenomeSpace REST-ful web interfaces from the Python programming language. (Full disclosure: this was my first attempt to use Python so I am sure there are stylistic issues and more elegant ways to do this). Below, we'll review the steps to connect to the GenomeSpace Data Manager and list the user's home directory in an interactive Python session. 1. Import the libraries we'll be using# set up imports import urllib2 import json 2. Set up a password manager basic authentication# set some properties gsUsername = 'ted' gsPassword = '*****' 3. Retrieve the GenomeSpace server URLs from the published web page
# get all the GenomeSpace URLs from the main web page
opener = urllib2.build_opener()
allUrls = opener.open('http://www.genomespace.org/sites/genomespacefiles/config/serverurl.properties')
urlMap = {}
for line in allUrls:
tokens = line.split("=")
if (len(tokens) == 2):
urlMap[tokens[0]] = tokens[1][:-1] #the trailing \n is not handled properly
idUrl = urlMap['prod.identityServerUrl']
dmUrl = urlMap['prod.dmServer'] + '/v1.0/'
4. Use basic authentication to get a gs-token to use for the rest of the callsauth_handler = urllib2.HTTPBasicAuthHandler() auth_handler.add_password(realm=" GenomeSpace ", uri=idUrl, user=gsUsername, passwd=gsPassword) opener = urllib2.build_opener(auth_handler) urllib2.install_opener(opener) urllib2.urlopen(idUrl) 5. Get the GSDirectory JSON object
req = urllib2.Request(dmUrl + 'file/Home/' + gsUsername)
req.add_header('Cookie','gs-token='+token)
mydir = sslopener.open(req)
homedirJson = mydir.read()
# turn it into a real object from a JSON string
homeDirObj = json.loads(homedirJson)
6. Get the URL for the first file and download its contents
firstUrl = homeDirObj['contents'][0]['url']
req2 = urllib2.Request(firstUrl)
req2.add_header('Cookie','gs-token='+token)
firstFileContents = sslopener.open(req2)
print firstFileContents
That's it! For more details on how to use the GenomeSpace REST-ful APIs, the structure of the JSON objects, etc., please refer to the documents on the Technical Documentation page.
|