Loading Data into GenomeSpace
|
Load a Gene List from Reactome Pathway into GenomeSpaceSummary This recipe provides an outline of how to use the Reactome pathway browser to idenity a list of genes or proteins in a pathway, then save the list as a file in your GenomeSpace data store. Input:
For more information about the Reactome pathway browser, see http://wiki.reactome.org/index.php/Usersguide#The_Pathway_Browser
Recipe Details 1. Click on the Reactome icon in the GenomeSpace toolbar
You will be sent to the Reactome website. 2. Click on Browse Pathways
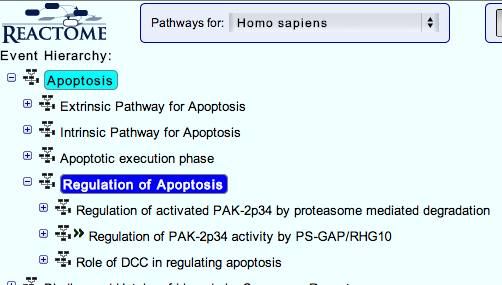
This will take you to the Reactome pathway browser.
3. You can navigate the pathway hierarchy by clicking on the
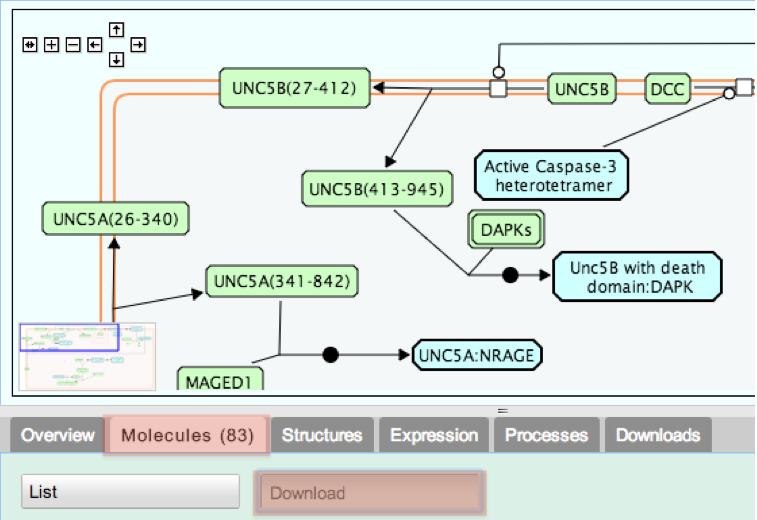
4. From the tabs below the pathway diagram, select Molecules, then click the Download button.
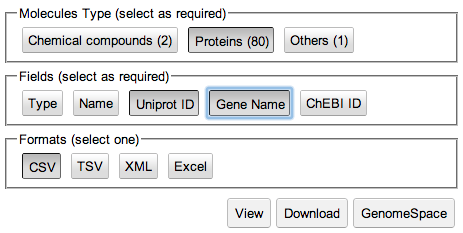
This takes you to the Download form. By default, all fields (buttons) are selected, as indicated by darker shading. 5. De-select all fields except Uniprot ID and Gene Name by clicking on each button each button that you do not want selected.
6. Click on View to preview the gene list.
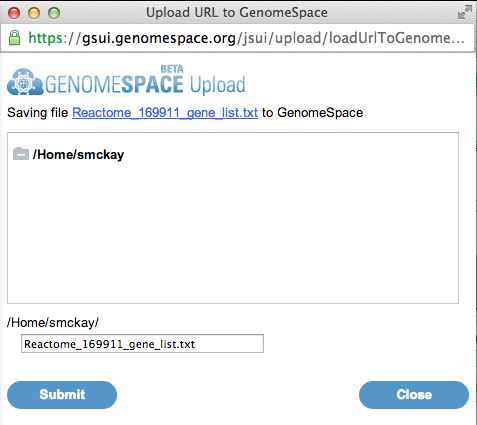
This shows a preview of the text that will be saved to a file. Make adjustments to your selection as required. 7. To save to your GenomeSpace data store, click the GenomeSpace button.
A window will pop up with a GenomeSpace dialog. 8. Select a file name and location to save the file to in your GenomeSpace data store. Click Submit to save the file, then click Close.
9. Return to the GenomeSpace website. Your gene list should now be there, after refreshing the page. |
|
<< Load Data from Synapse to GenomeSpace | Up | Pathway Enrichment Analysis with Reactome >> | |



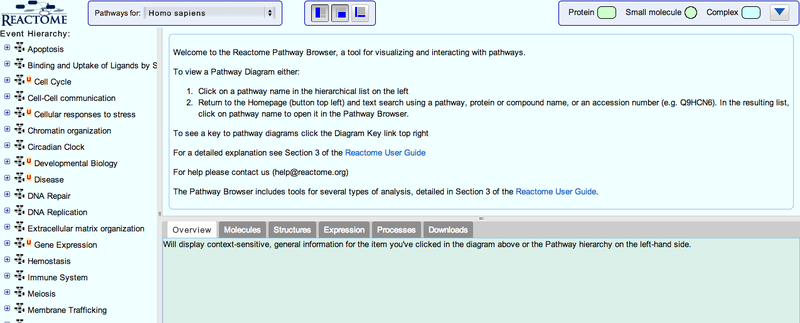
 symbol on the left side of the pathway labels. Open Apoptosis->Regulation of Apoptosis
symbol on the left side of the pathway labels. Open Apoptosis->Regulation of Apoptosis