Loading Data into GenomeSpace
|
Load Data from UCSC Genome Browser to GenomeSpace
1. Launch UCSC Table Browser from GenomeSpace.
Click the UCSC Table Browser icon in the Data Sources toolbar to launch it.
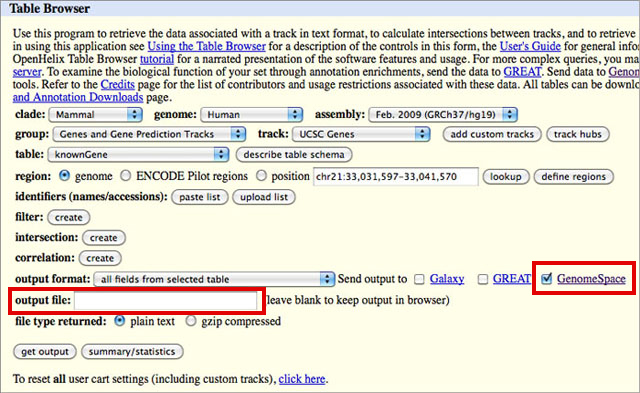
2. Locate data in UCSC Table Browser and send to GenomeSpace.
NOTE: You must add the correct extension to your file name. If, for instance, you select BED in the output format drop-down, you will need to add .bed to your output file name. The UCSC Table Browser does not do this for you, and some of the GenomeSpace tools depend on the file extension to determine file type.
|
|
<< Load GEO Data from InSilico DB to GenomeSpace | Up | Load Data from Synapse to GenomeSpace >> | |