Loading Data into GenomeSpace
|
Pathway Enrichment Analysis with ReactomeSummaryThis recipe provides an outline of how to perform pathway enrichment analysis on the reactome website using a list of genes or protein Ids in GenomeSpace. Given a list of gene ideintifers, the goal is to query the reactome database for pathways whose components include the proteins in the supplied list. Enriched pathways are then displayed with the specified proteins highlighted, and the diagrams can be saved back to GenomeSpace. Input:
Input Formats: A list of NCBI/Entrez gene ids: 2 21 10257 8038 ... A list of UniProt ids:
O00139 Tab-delimited expression data:
#Probeset 10h_control 10h 14h 18h 24h For more information about input data, see http://wiki.reactome.org/index.php/Usersguide#Gene_list_Dataset
Recipe Details 1. Drag your gene list file onto the Reactome OR click the icon and select Launch on File, then select your gene list file
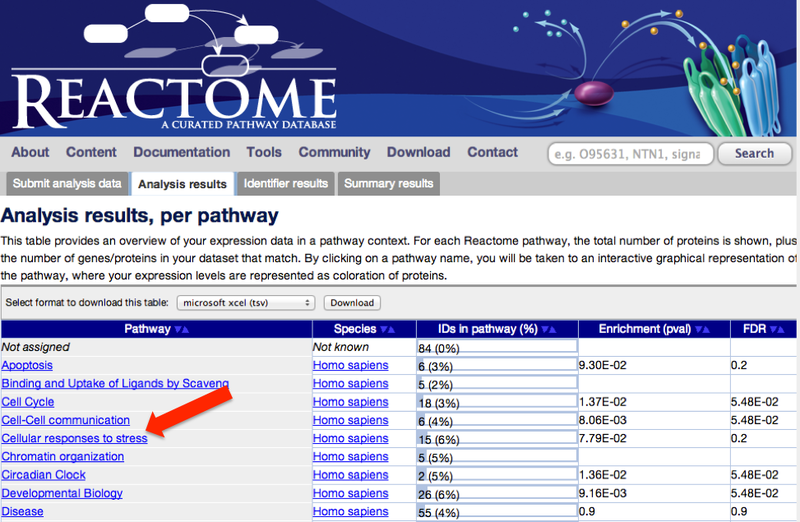
You will be sent to the Reactome website. 2. When the analysis is complete, click on one of the top-level pathways, at the left hand side of the screen.
This will take you to the Reactome pathway browser.
3. Navigate the pathway hierarchy by clicking on the
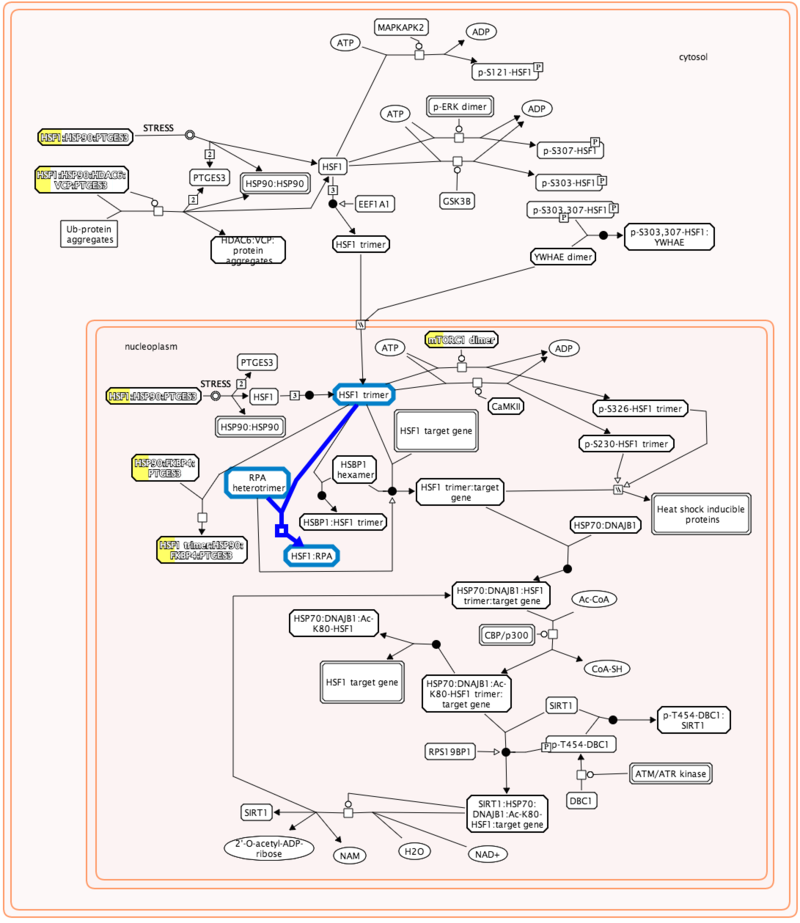
The pathway diagrams are colorized according to the number of enriched proteins.
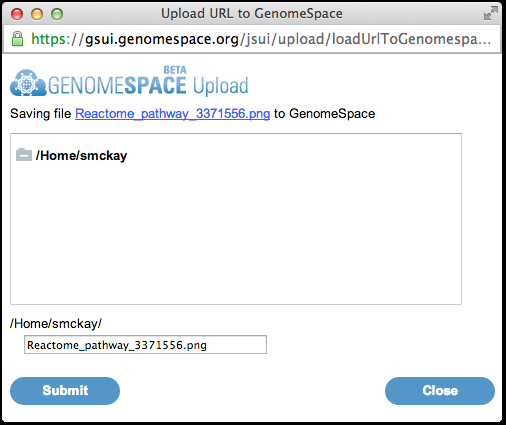
4. Once you have selected your pathway, click the
A window will pop up with a GenomeSpace dialog. 5. Select a location to save the file to in your GenomeSpace data store. Click Submit to save the file, then Close.
6. Return to the GenomeSpace website. Your pathway diagram file should now be there, after refreshing the page.
|
|
<< Load a Gene List from Reactome Pathway into GenomeSpace | Up | >> | |


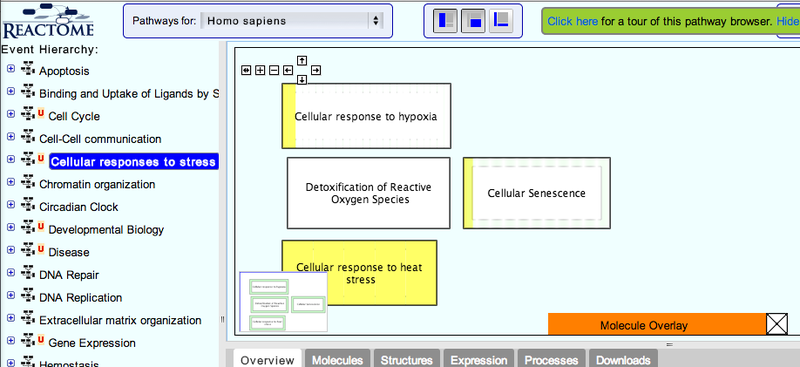
 symbol on the left side of the pathway labels.
symbol on the left side of the pathway labels.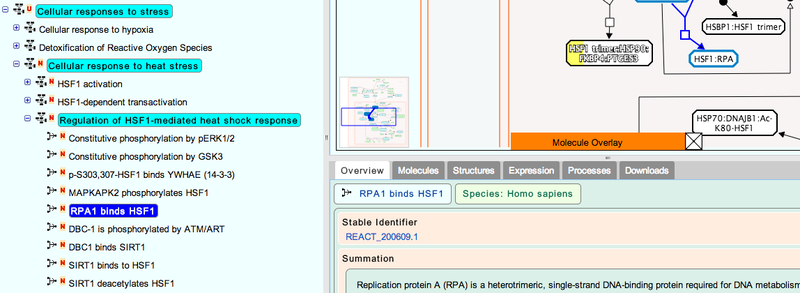
 icon and select Save Diagram to GenomeSpace.
icon and select Save Diagram to GenomeSpace.