Loading Data into GenomeSpace
|
Load Data from ArrayExpress to GenomeSpace
1. Launch ArrayExpress from GenomeSpace.
Click the ArrayExpress icon in the toolbar to launch ArrayExpress.
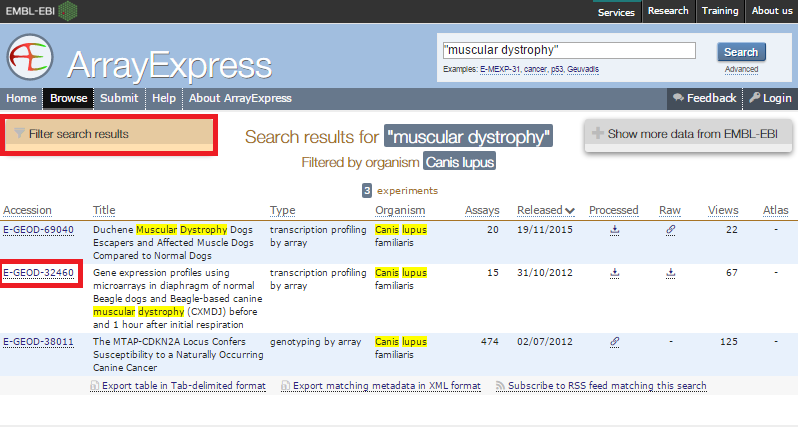
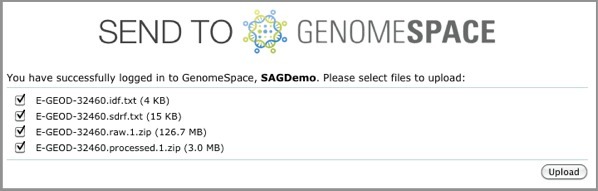
2. Locate data in ArrayExpress and send to GenomeSpace.
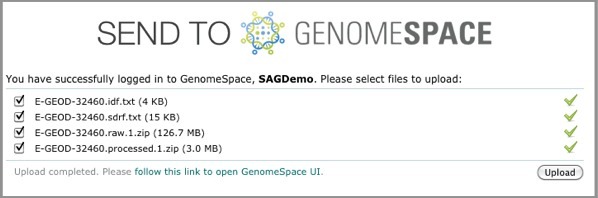
ArrayExpress shows you the upload status of each file.
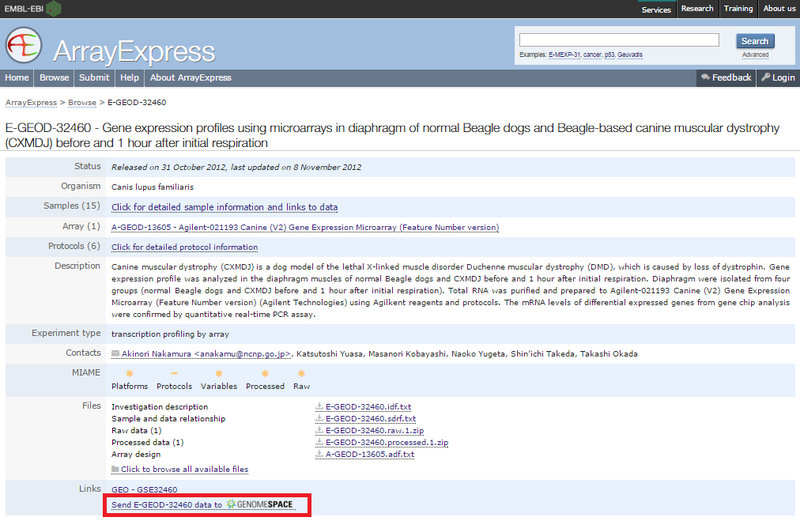
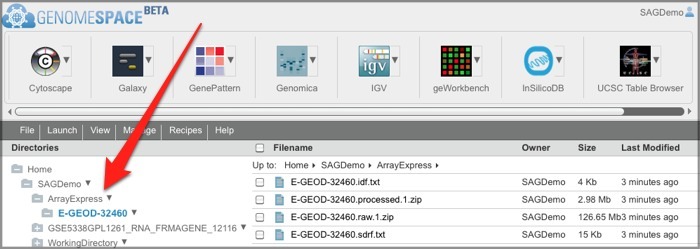
3. Locate your ArrayExpress data in GenomeSpace.If this is your first time loading ArrayExpress data into GenomeSpace, ArrayExpress creates an ArrayExpress folder in the top level of your GenomeSpace home directory. Subsequently, ArrayExpress creates a separate subdirectory in that folder for each experiment you upload.
One way you can use the ArrayExpress experiment files (which are in MAGE-TAB format) you download is to send them to GenePattern's MAGETABImportViewer module to create GCT and CLS files for analysis. See the MAGETABImportViewer module documentation for more information on the specific MAGE-TAB format the module can receive. |
|
<< Upload Data to GenomeSpace | Up | Load Data from the cBioPortal to GenomeSpace >> | |