Tool Guide |
Cytoscape 3Project Website: http://www.cytoscape.org New Documentation for Cytoscape 3 is on its way... In the meantime, below is the help from the previous revision (Cytoscape 2.8) Cytoscape is an open-source bioinformatics software platform for visualizing molecular interaction networks and biological pathways, and integrating these networks with annotations, gene expression profiles, and other state data. Cytoscape core distribution provides a basic set of features for data integration and visualization. Additional features are available as plugins. Cytoscape is a Java-based application that downloads to your local machine when you launch it from GenomeSpace. Once it's downloaded to your machine (after a launch from GenomeSpace), you can start it locally (by opening the JNLP) or launch from GenomeSpace. If you launch Cytoscape from the Cytoscape website, at this time it will not be the version that is linked to GenomeSpace. Like several other applications in the GenomeSpace suite of tools, Cytoscape can be expanded with plug-ins. You can download plug-ins for your Cytoscape by choosing Plugins>Manage Plugins. Cytoscape has several GenomeSpace menu items. Logging InWhen you launch Cytoscape from GenomeSpace, your login should be handled seamlessly. If you start from your GenomeSpace-enabled JNLP of Cytoscape and want to log into GenomeSpace in order to access your Amazon cloud storage or other GenomeSpace tools, you will be asked to log in with your GenomeSpace username and password.
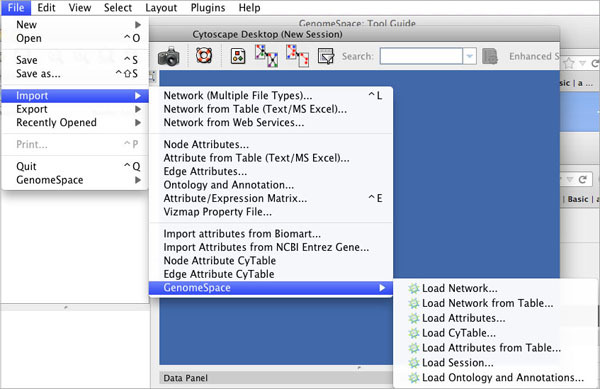
You can also Reset Password on your existing GenomeSpace account. Import MenuClicking File>Import>GenomeSpace provides several options for importing files from GenomeSpace.
|
||||||||||||||||||||||||||||||
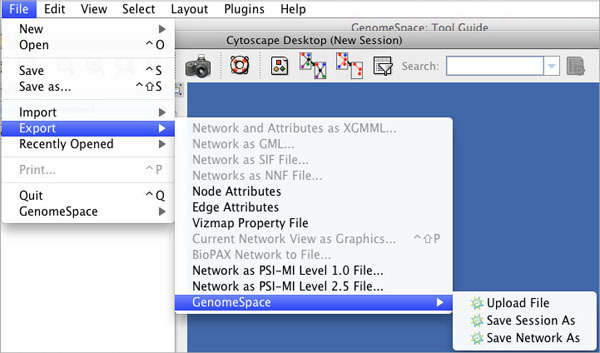
| Upload File | Uploads a file from your local machine to the root level of your GenomeSpace cloud storage. |
| Save Session As | Saves a session file (CYS), containing all networks, attributes (for node/edge/network), Desktop states (selected/hidden nodes and edges, window sizes), Properties, some plugin states, and Visual Styles in your present working state to your GenomeSpace cloud storage. |
| Save Network As | Save a network to your GenomeSpace cloud storage. |
GenomeSpace Menu
Clicking File>GenomeSpace provides several options for managing your files in your GenomeSpace cloud storage and using GenomeSpace to launch other applications.
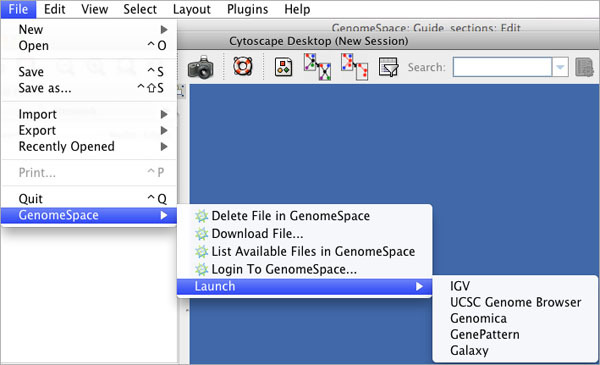
| Delete File in GenomeSpace | Delete a file from your GenomeSpace cloud storage. |
| Download File | Download a file in your GenomeSpace cloud storage to your local machine. |
| List Available Files in GenomeSpace | Navigate your GenomeSpace directories. |
| Login To GenomeSpace | Log into your GenomeSpace account. |
| Launch | Launch another GenomeSpace tool |