Tool Guide |
TrinityThe Trinity Cancer Transcriptome Analysis Toolkit (CTAT) aims to provide tools for leveraging RNA-Seq to gain insights into the biology of cancer transcriptomes. Bioinformatics tool support is provided for mutation detection, fusion transcript identification, and de novo transcript assembly. Trinity CTAT was developed at The Broad Institute and is hosted on the Trinity CTAT Galaxy portal by the National Center for Genome Analysis Support (NCGAS) at Indiana University. CTAT is funded by the National Cancer Institute Informatics Technology for Cancer Research (NCI ITCR) program. More information about Trinity CTAT can be found using the following links:
Creating an AccountBefore you can launch Trinity from GenomeSpace, you must first sign up for a Trinity account through the National Center for Genome Analysis Support (NCGAS). This can be accomplished by filling out the Trinity Galaxy Registration Form. Trinity Galaxy support by NCGAS is publically funded, so users are required to sign up and describe the nature of their work and their sources of funding. If you have any questions about this form, please contact the NCGAS team at help@ncgas.org. Once you have submitted your registration form, you should receive an email from the NCGAS team with your account information and temporary password. This email should arrive within 24 hours of submitting your application.
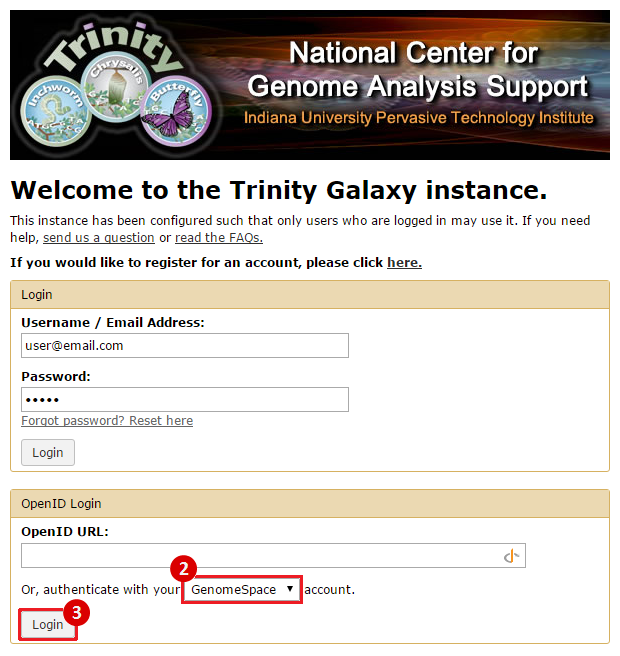
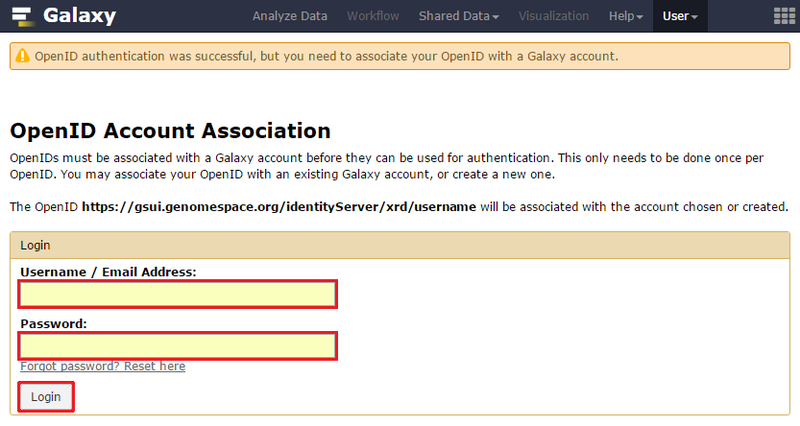
Connecting Your AccountOnce you have signed up for and activated your Trinity CTAT account, you can connect your Trinity account to GenomeSpace using the following method:
Once you have connected your account, you can begin using Trinity normally.
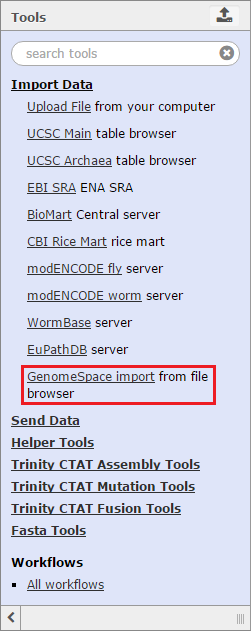
Trinity ToolsTrinity encompasses a suite of tools hosted on a Galaxy server. There are three types of tools: (1) tools for interacting with GenomeSpace; (2) auxiliary tools that are common to Galaxy builds; and, (3) Trinity-specific tools which perform different functions or analyses. Get DataTo import data to Trinity from GenomeSpace, click Get Data in the Tools menu and select GenomeSpace import from file browser. This opens a view of your GenomeSpace directories. The navigation is a little different here than it is in GenomeSpace, since there is no separate directories pane. Double-click folders to open them, and double-click ..(To Parent) to return to the previous level of the folder structure.
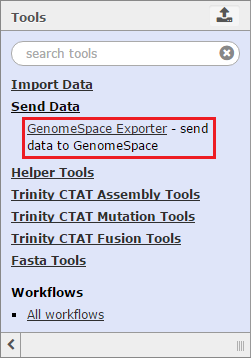
Select the checkboxes for the files you want to bring into Trinity. You can optionally select a file format from the file's drop-down menu, provided there is a converter. Click the Send to Galaxy button to queue the files for import into Trinity. Send DataTo send data back to GenomeSpace from Trinity, click Send Data in the Tools menu and select GenomeSpace Exporter.
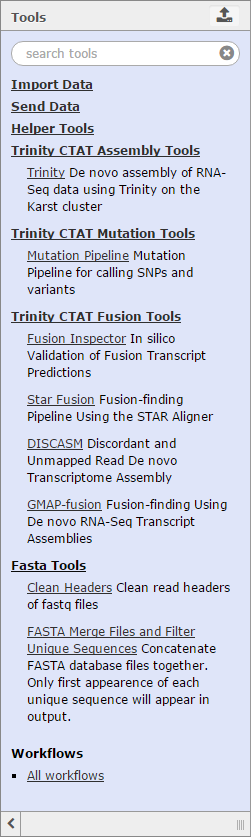
Other Galaxy ToolsTrinity also has built-in Galaxy tools to help with basic file manipulation and other functions. These tools can by found by clicking on Helper Tools in the Tools menu.
To learn about other Galaxy tools that can be found in Trinity, please visit the Galaxy Wiki Resource. Trinity-Specific ToolsThe Trinity CTAT suite of tools is specific to this build of Galaxy, and is not currently deployed on the main Galaxy server which is also connected to GenomeSpace. Currently, CTAT supports tools and functions for completing analyses such as:
More information about the capabilities of these specific tools can be found on the Trinity CTAT wiki page. In the future, Trinity CTAT may also support lincRNA classification, foreign transcript detection, and single-cell cancer transcriptome analysis.
|
|
<< Tool Term Glossary | Up | >> | |