Tool Guide |
FireBrowseFireBrowse is a companion portal to the Broad Institute GDAC Firehose analysis pipeline, and was developed to cull and analyze data generated by The Cancer Genome Atlas (TCGA), which characterizes and identifies genomic patterns in human cancer models. Backed by a powerful compute infrastructure, programming interface, online reports and modern graphical tools, FireBrowse provides a simple yet capable means of visually and programmatically exploring one of the most comprehensive and deeply-characterized open cancer datasets in the world. FireBrowse provides access to a variety of cancer genomics data, such as clinical annotations, DNA copy number, miR, miRseq, mRNA and mRNAseq; as well as a comprehensive suite of more than 100 interdependent analyses of those data, including: correlations, clustering, and GISTIC and MutSigCV. FireBrowse is developed at the Broad Institute of MIT and Harvard. More information about FireBrowse can be found using the following links:
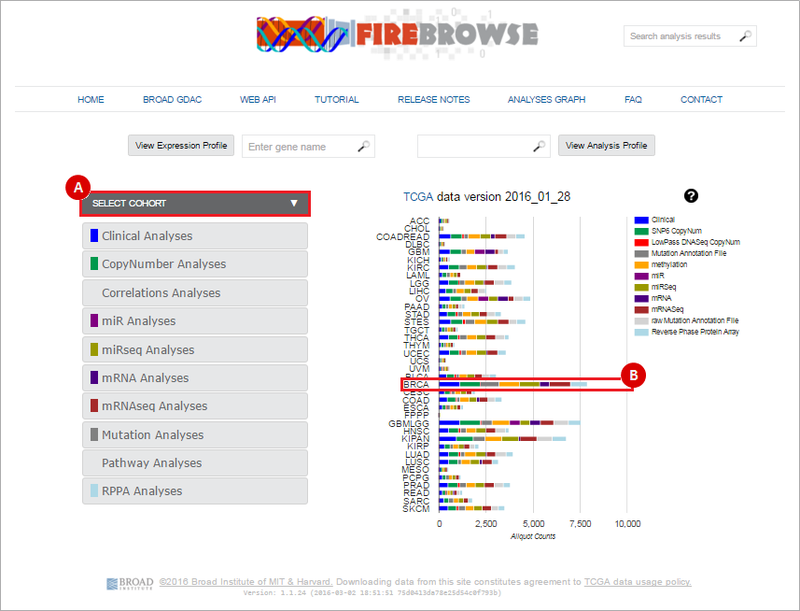
Downloading data from FireBrowse to GenomeSpaceWhen you launch FireBrowse from GenomeSpace, you will be automatically redirected to the FireBrowse front page (http://firebrowse.org/). On this page, you can access FireBrowse data by selecting a cohort. To do this, you can (A) select a cohort from the drop-down menu; or, (B) simply click on the respective row of the samples barchart for that cohort. In the example below, we will be downloading Breast invasive carcinoma (BRCA) data. Therefore, we can either: (A) “Breast invasive carcinoma (BRCA)” from the drop-down menu; or (B) click on the BRCA barchart.
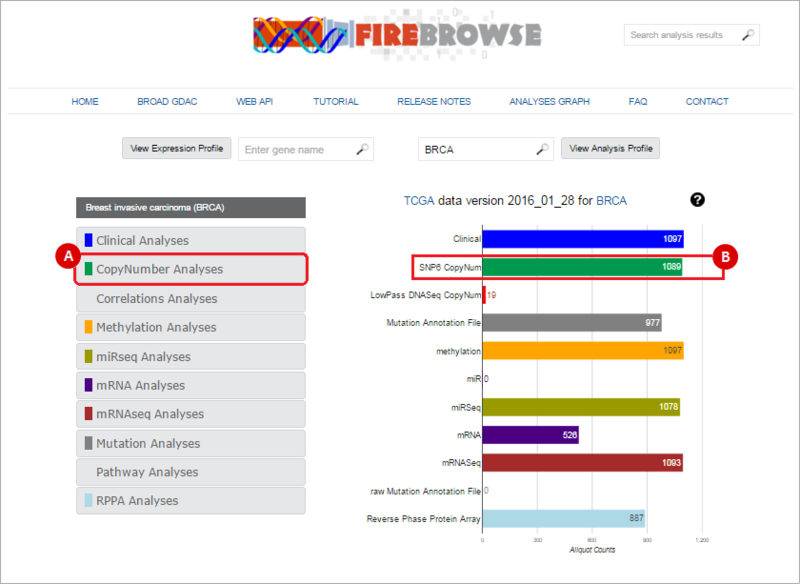
Next, select the data that you wish to download. FireBrowse collects data from a variety of analyses: clinical, copy number, miR, miRseq, mRNA, mRNAseq, mutation and pathway analyses. To select a data type, either: (A) select the data type from the left-hand menu; or (B) click on the corresponding barchart. In our example, we will download SNP6 Copy Number variation data. We therefore either: (A) Click on the "CopyNumber Analyses" box; or (B) click on the "SNP6 CopyNum" barchart.
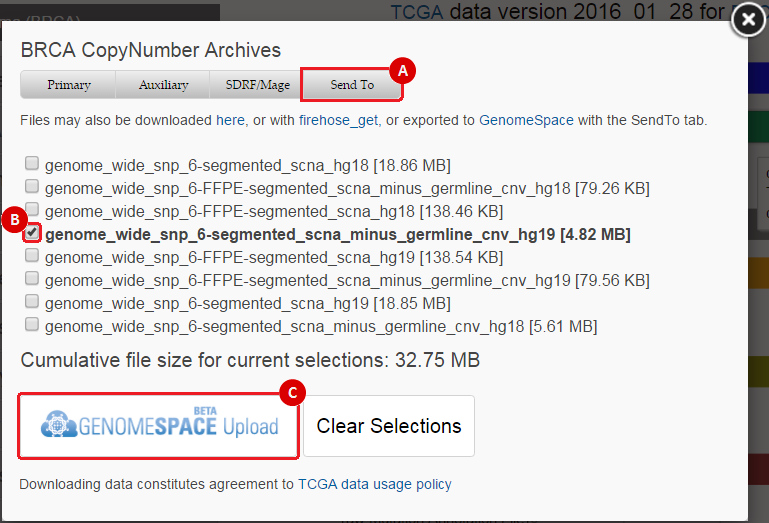
When you select a data type to download, this will prompt a data download interface. To send data to GenomeSpace, use the following method:
This will prompt you to choose a GenomeSpace directory to save the file(s) to. Once you have chosen a directory, you can click the "Submit" button. This will send the file(s) to your GenomeSpace directory as a .tar.gz file. When you return to GenomeSpace, you will have to right-click on the file and choose "Expand Archive". This will unzip the .tar.gz file and create a new folder with the same name. The files you chose to download in Step B will be available in that folder. |