Tool Guide |
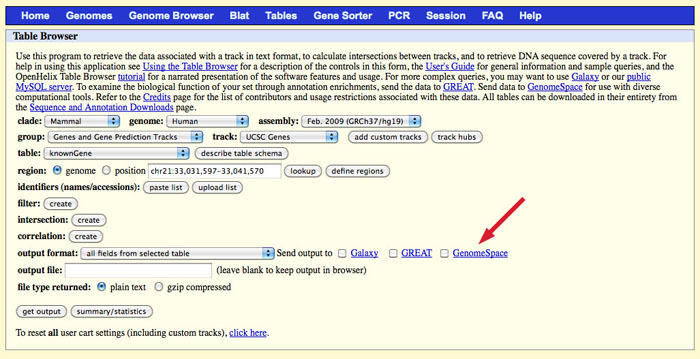
UCSC Table BrowserSending Files from the UCSC Table Browser to GenomeSpaceProject Website: http://genome.ucsc.edu/index.html The UCSC Table Browser is a valuable data source linked to GenomeSpace. Since it is a data source, you cannot launch the UCSC Table Browser on a file in GenomeSpace. You can only launch the UCSC Table Browser from its button in GenomeSpace, and then send files from the UCSC Table Browser to GenomeSpace. The Table Browser allows you to retrieve data associated with a track in text format, to calculate intersections between tracks, and to retrieve DNA sequence covered by a track. After you select the options for your output file, you can opt to send your output file to your GenomeSpace cloud storage. For help using the Table Browser's options, see the UCSC Table Browser documentation.
To output your selected track to GenomeSpace, remember to select the checkbox for GenomeSpace. Select an output format. In the output file field, you need to specify the name of your output file. There are two options for this:
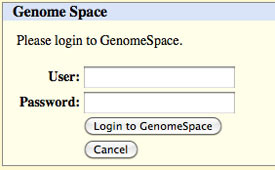
NOTE: You must add the correct extension to your file name. If, for instance, you select BED in the output format drop-down, you will need to add .bed to your output file name. The UCSC Table Browser does not do this for you, and some of the GenomeSpace tools depend on the file extension to determine file type. When you click get output, you will be asked to log into GenomeSpace, if you have not previously logged in via the UCSC Genome Browser.
After you log in (or if you previously logged in), you will see an uploading message.
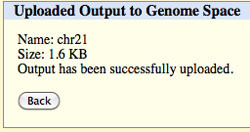
Followed by a completion message.
Click Back to return to the Table Browser. The file you sent to GenomeSpace should appear in your root directory.
Read more documentation on how to load data from UCSC Genome Browser to GenomeSpace. |