Galaxy
Project Website: http://galaxyproject.org/
Galaxy is an open-source, scalable framework for tool integration that allows users to analyze multiple alignments, compare genomic annotations, and profile metagenomic samples, among many possible analyses; workflows allow the linking together of analyses.
Please note that during the GenomeSpace Beta, you will be connected with a Galaxy test server.
Logging In
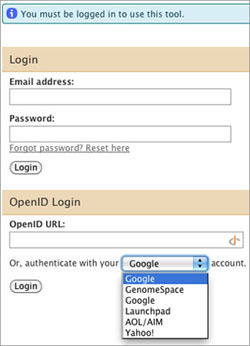
When you launch Galaxy from GenomeSpace, you will see a login window. Do not enter anything in the Email address, Password, or OpenID URL fields!
In the final line under the OpenID login section ("Or, authenticate with your _____ account."), select GenomeSpace and click Login.
Galaxy Tools
Galaxy has its own tools that perform different functions or analyses, and there are tools for interacting with GenomeSpace.
Get Data
To import data to Galaxy from GenomeSpace, click Get Data in the Tools menu. Under that, select GenomeSpace import from file browser. This opens your GenomeSpace directories. The navigation is a little different here than it is in GenomeSpace, since there is no separate directories pane. Double-click folders to open them, and double-click ..(To Parent) to return to the previous level of the folder structure.
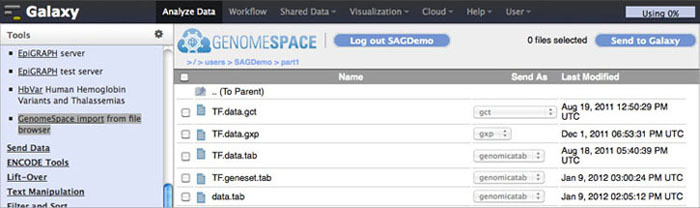
Select the checkboxes for the files you want to bring into Galaxy. You can then select a file format from the file's drop-down menu, if there is a converter. Click the Send to Galaxy button to queue the files for import into Galaxy.
Send Data
To send data back to GenomeSpace from Galaxy, click Send Data in the Tools menu. Under that, select GenomeSpace Exporter.
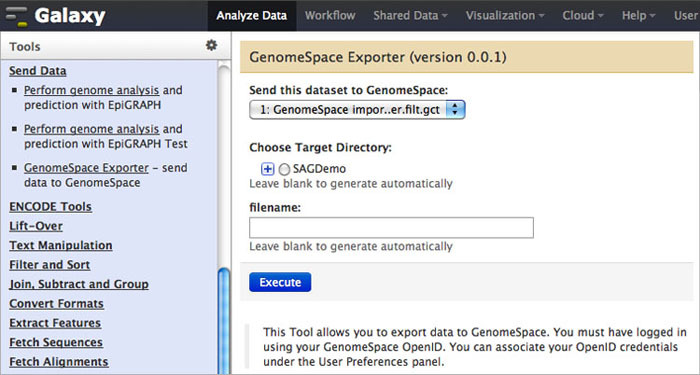
-
Select the dataset you want to export from the drop-down menu.
-
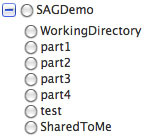
Select the target directory -- you can open the base directory by clicking the
 so that you can select a subdirectory: so that you can select a subdirectory:
-
You can enter a file name. If you do not enter one, Galaxy will generate a file name automatically.
-
Click Execute to send the file to GenomeSpace.
Learn how to send data from Galaxy to IGV using GenomeSpace.
|




